Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Investigating non-specific binding to chemically engineered sensor surfaces using liposomes as models†
C.
Fenzl
a,
C.
Genslein
a,
C.
Domonkos
b,
K. A.
Edwards
c,
T.
Hirsch
a and
A. J.
Baeumner
*ac
aInstitute of Analytical Chemistry, Chemo- and Biosensors, University of Regensburg, Universitaetsstr. 31, 93053 Regensburg, Germany. E-mail: antje.baeumner@ur.de
bResearch Centre for Natural Sciences, Hungarian Academy of Sciences, Magyar tudósok körújta 2, 1117, Budapest, Hungary
cDepartment of Biological and Environmental Engineering, Cornell University, Riley-Robb Hall, 111 Wing Drive, Ithaca, NY 14853-5701, USA
First published on 15th June 2016
Abstract
Nanoparticles are ubiquitously used for signal enhancement in (bio)sensors, but their true possible performance is typically hampered by non-specific binding. A better understanding of the nature and the prevention of non-specific binding through surface engineering of the particles and sensor surfaces is needed to intelligently design (bio)sensors and potentially avoid bulk blocking methods. Hence, two types of liposomes were used as model for signal-enhancing nanoparticles. Their surface was engineered to bear negative surface charge. One type was synthesized with additional 6 mol% –COOH groups. Their interaction with four typical chemically modified sensor surfaces was then mechanistically characterized by surface plasmon resonance (SPR) spectroscopy. It was shown that the non-specific binding can be described with Langmuir isotherms providing quantitative information of dissociation constants and surface loading with especially high correlation coefficients (>0.97) for all the studied sensor surfaces modified with hydrophilic alkane thiols. By tailoring the sensor surface chemistry, non-specific binding was significantly minimized. Here, carboxyl- or methyl-terminated surfaces performed best. In fact, the pairing of –COOH groups on the sensor surface with –COOH groups on the liposomes almost completely eliminated non-specific binding, resulting in a SPR signal change of only 1 mRIU (refractive index unit) at 100 μM phospholipid concentration. Surprisingly though, –OH groups on the surface, which are also commonly used in sensing applications, did not lead to decreased adsorption, but caused significant signal changes (4 mRIU at 100 μM phospholipid) due to non-specific binding. Overall, the mechanistic studies presented here demonstrate that by careful design of the nanoparticle surface and by choosing sensor surfaces with terminal –CH3 or –COOH groups, improved sensing (micro)systems with very low non-specific adsorption can be obtained.
Introduction
Sensor performance with respect to its limit of detection depends on a variety of factors that are independent of the inherent transducer and (bio)recognition's ability to provide highly sensitive signals. These factors include non-specific binding, the required surface protection via membranes or coatings that affect diffusion, signal enhancement capabilities, and specificity toward the analyte, among others. Non-specific binding may be the most limiting factor in microsystems with large surface-to-volume ratios, such as microelectrodes and microfluidic devices, as high background signals will obviate any advantages obtained through sensor sensitivity and signal enhancement. Especially the signal enhancement is affected by the negative effects of non-specific binding, as also background signals will be enhanced. Key should thus be the design of sensor surfaces that eliminate this type of non-specific interaction without the need of additional blocking reagents. Liposomes are very powerful means as signalling labels in the analytical sciences,1–4e.g. biosensors5 as well as in drug delivery.6–8 Their wide applications and the direly missing thorough investigations of non-specific binding to typical (bio)sensor surfaces make them the ideal model for the mechanistic studies presented here. Liposomes are artificial nanoscale vesicles consisting of a hydrophobic lipid bilayer that separates the hydrophilic inner cavity from the outer medium.5,9 Recently, Edwards et al.10 showed magnetic fluorescent liposomes to be very powerful signal amplifiers in heterogeneous binding assay formats where DNA-tagged liposomes were capable of enhancing the sensitivity and reducing assay times simultaneously. Furthermore, vesicles encapsulating quantum dots11 as well as liposomes incorporating high binding biorecognition elements such as ganglioside receptors12,13 enable attomolar detection of DNA without prior target amplification. Additionally, liposomes with high refractive index encapsulants have been demonstrated to significantly enhance the signal in surface plasmon resonance (SPR) spectroscopy.14Despite the high performance of liposome-based sensors and drug delivery systems, the dynamics of the interactions between the utilized vesicles and their target surfaces have only been marginally studied and are still not well understood. In particular, in microfluidic devices – a growing field in (bio)analytical research – high surface areas are encountered that can lead to increasing non-specific adsorption15 and need to be avoided by clever surface engineering strategies. Here, mechanistic understanding of the binding processes is crucial for the optimization of sensors as well as therapeutic applications based on liposomes, where intensive efforts in liposome engineering are made to maximize the uptake of a drug for the desired target.7 Therefore, it is of high importance to know how much, how fast and how strong the vesicles bind to the target surface specifically and non-specifically.
Liang et al.16 investigated the effect of flow rate and water content on targeted liposome interactions via surface plasmon resonance (SPR) spectroscopy and quartz crystal microbalance (QCM) studies. They found that increasing the flow rate decreases the maximum amount of bound liposomes and the equilibrium constant, as does decreasing the water content in the bound vesicle layer. Further, the morphology of the lipid layer on a surface can be controlled by the surface chemistry on the substrate.17 Granqvist et al.17 showed that low-molecular-weight dextran-based surfaces facilitate the formation of supported lipid bilayers, whereas polyethylene glycol-based thiol-surfaces lead to supported vesicular layers. These findings are of high importance when using liposomes as carriers for drugs6,18 or signal molecules10 that should be released in a controlled manner. Recently, Calver et al.19 were able to monitor lipid membrane–surface interactions such as liposome adsorption and deformation using single-particle fluorescence of conjugated poly[5-methoxy-2-(3-sulfopropoxy)-1,4-phenylene-vinylene] on SiO2 nanoparticles.
Extensive studies of lipid vesicle adsorption and supported lipid bilayer formation have been performed by the groups of Reviakine,20,21 Richter and Brisson,22,23 and Kasemo.24–26 They studied the behaviour of liposomes on sensor substrates such as SiO2,24–26 TiO2,20,25 gold24–26 or mica22,23 that contributed significantly to the general understanding of liposome binding to standard sensor substrates of SPR, QCM and atomic force microscopy and the conditions for bilayer formation.
However, again, studies are missing that investigate binding interactions with the predominant surface modifications applied in (bio)analytical assays and sensors, e.g. surfaces modified with terminal carboxyl or hydroxyl groups.14,27 Yet, this knowledge will lead to viable surface engineering strategies that contribute to higher selectivity and reusability, as the commonly used bulk blocking with proteins or polymers is often not sufficient enough to prevent non-specific binding completely. In addition, the liposome formulations used in the previous studies differ clearly from those used in high-performance sensing approaches.1,14,28 As the composition influences sensing performance it must also be used when trying to understand and thereby minimize non-specific binding.
Here, we present the systematic study of non-specific binding interactions between highly stable anionic liposomes and surfaces with varying surface charge and hydrophilicity (Fig. 1). We focus on this type of liposome, as it is favourably used in high-performance sensing applications1,10 that are often combined with microfluidic sample handling.15,29 Liposomes with negative surface charge have been shown not to rupture as easily as positively charged ones when adsorbing to solid surfaces.30 Also, with most biological molecules bearing negative charges, non-specific binding of anionic liposomes favours their use in bioassays. The vesicles were characterized in close detail regarding size and surface charge. The binding studies were performed using SPR spectroscopy that has been shown to be the ideal tool for label-free monitoring of such interactions in real-time resolution.16,17,31 Further, the influence of various temperatures on the binding behaviour was analysed. With our present work, we will broaden the knowledge on liposome–solid surface interactions; this will benefit a wide range of applications by providing clever surface engineering strategies preventing non-specific adsorption without the need of bulk blocking or surface coating.
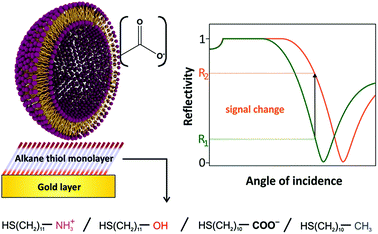 | ||
| Fig. 1 Sensing scheme for the interactions between anionic liposomes and different self-assembled monolayers on gold. High non-specific binding will result in high SPR signal changes. | ||
Experimental
Materials
1,2-Dipalmitoyl-sn-glycero-3-phosphocholine, 1,2-dipalmitoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] sodium salt, N-glutaryl-1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine, and the extrusion membranes as well as the extrusion kit were purchased from Avanti Polar Lipids (Alabaster, AL, USA). 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), sodium azide, cholesterol, phosphotungstic acid (PTA), 11-mercaptoundecanoic acid, 11-mercaptoundecanol, and 1-mercaptoundecane were obtained from Sigma-Aldrich (Taufkirchen, Germany). 11-Mercaptoundecyl amine hydrochloride was purchased from ProChimia Surfaces (Sopot, Poland). All other chemicals used in these experiments were ordered from VWR (Darmstadt, Germany).Preparation of liposomes
The liposomes were prepared according to a slightly modified protocol developed by Edwards et al.32 DPPC, DPPG and cholesterol (40.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20.1
20.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 51.7 μmol, respectively) for the untagged liposomes, and DPPC, DPPG, cholesterol and N-glutaryl-DPPE (40.9
51.7 μmol, respectively) for the untagged liposomes, and DPPC, DPPG, cholesterol and N-glutaryl-DPPE (40.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20.1
20.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 51.7
51.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.3 μmol, respectively) for the liposomes with additional –COOH tags were dissolved in an organic solvent mixture consisting of 3 mL chloroform and 0.5 mL methanol.
7.3 μmol, respectively) for the liposomes with additional –COOH tags were dissolved in an organic solvent mixture consisting of 3 mL chloroform and 0.5 mL methanol.
Then, the solution was sonicated for 1 min in a sonication bath (Bandelin Sonorex Digitec DT 255 H) at 45 °C for homogeneous mixing. A 45 °C salt solution (2 mL of 300 mmol L−1 NaCl) was added to the lipid mixture and then again sonicated for 4 min. The organic solvent was removed at 45 °C and 380 mbar for 20 min using a rotary evaporator. The mixture was vortexed before and after a second addition of 2 mL 45 °C 300 mmol L−1 NaCl. The flask was returned to the rotary evaporator for 20 min at 45 °C and 380 mbar and then for 20 min at 280 mbar. The crude liposome dispersion was further extruded at 50 °C 21 times through 1.0 μm Nucleopore membranes (Whatman, Florham Park, NJ, USA), followed by 21 times through 0.4 μm membranes. The liposomes were purified via size-exclusion chromatography with a Sephadex G-50 in a 15 × 1.6 cm column at ∼4 mL min−1 using HEPES-saline-sucrose buffer (HSS; 10 mmol L−1 HEPES, 200 mmol L−1 sodium chloride, 200 mmol L−1 sucrose, 1.5 mmol L−1 sodium azide at pH 7.5). The fractions containing liposomes were combined and dialyzed overnight against HSS before storage at 4 °C.
Liposome characterization
For transmission electron microscopy, the purified liposome dispersions were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in HEPES-saline buffer (10 mmol L−1 HEPES, 200 mmol L−1 sodium chloride, 1.5 mmol L−1 sodium azide at pH 7.5). A carbon-coated copper grid was covered with a 2 μL drop of the suspension for 90 s. After removing the excess liposomes by washing with 5 μL of 8.7 mmol L−1 PTA aqueous solution, the vesicles left on the TEM-grid were negatively stained33 with a 5 μL drop of 8.7 mmol L−1 PTA aqueous solution for 30 s in order to enhance the TEM contrast. The excess staining solution was removed with a filter paper. Transmission electron micrographs were acquired with a transmission electron microscope (Philips CM 12). Dynamic light scattering measurements were performed with the same liposome dispersions (1
10 in HEPES-saline buffer (10 mmol L−1 HEPES, 200 mmol L−1 sodium chloride, 1.5 mmol L−1 sodium azide at pH 7.5). A carbon-coated copper grid was covered with a 2 μL drop of the suspension for 90 s. After removing the excess liposomes by washing with 5 μL of 8.7 mmol L−1 PTA aqueous solution, the vesicles left on the TEM-grid were negatively stained33 with a 5 μL drop of 8.7 mmol L−1 PTA aqueous solution for 30 s in order to enhance the TEM contrast. The excess staining solution was removed with a filter paper. Transmission electron micrographs were acquired with a transmission electron microscope (Philips CM 12). Dynamic light scattering measurements were performed with the same liposome dispersions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in HEPES-saline buffer). A disposable polystyrene cuvette was filled with 1 mL of the suspension and analysed with the particle sizer in the backscattering mode at an angle of 173° (Malvern Zetasizer nano series) at 25 °C after an equilibration time of 120 s. The autocorrelation of the intensity recorded over time is related to the geometry of the liposomes under observation. After 30 consecutive measurements, the mean hydrodynamic radius and a polydispersity index were extracted from the autocorrelation data.
100 dilution in HEPES-saline buffer). A disposable polystyrene cuvette was filled with 1 mL of the suspension and analysed with the particle sizer in the backscattering mode at an angle of 173° (Malvern Zetasizer nano series) at 25 °C after an equilibration time of 120 s. The autocorrelation of the intensity recorded over time is related to the geometry of the liposomes under observation. After 30 consecutive measurements, the mean hydrodynamic radius and a polydispersity index were extracted from the autocorrelation data.
The electrophoretic mobility of the liposomes was measured with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution in HEPES-saline buffer. A Folded Capillary Cell (Malvern DTS1070) was charged with approximately 800 μL of the liposome dispersions and equilibrated to 25 °C for 120 s. The mean electrophoretic mobility was determined by laser Doppler velocimetry with the Zetasizer nano series. The Smoluchowski model was employed to determine the zeta potential of the dispersions.34
100 dilution in HEPES-saline buffer. A Folded Capillary Cell (Malvern DTS1070) was charged with approximately 800 μL of the liposome dispersions and equilibrated to 25 °C for 120 s. The mean electrophoretic mobility was determined by laser Doppler velocimetry with the Zetasizer nano series. The Smoluchowski model was employed to determine the zeta potential of the dispersions.34
For the determination of the phospholipid concentration of liposomes, inductively coupled plasma-atomic emission spectroscopy (ICP-AES) was used. A volume of 20 μL of liposomes was diluted in HNO3 (c = 0.5 mol L−1) to a total volume of 3 mL. The mixture was vortexed and sonicated thoroughly. Phosphorus standard solutions were prepared with concentrations of 1, 5, 10, 25, 50 and 100 μmol L−1 phosphate in 0.5 mol L−1 HNO3 for calibration. The phosphorus content was measured via ICP-AES (Spectro Flame-EOP, Analytical Instruments GmbH, Kleve, Germany) at the phosphorus specific wavelength of 178.29 nm.
Characterization of the formation of a self-assembled monolayer on gold
Cyclic voltammograms of gold electrodes (0.37 mm2) were recorded in HEPES buffer with 5 mM K3[Fe(CN)6] from −0.3 V to 0.6 V against a Ag/AgCl reference electrode (scan rate = 100 mV s−1) with an electrochemical analyser (CH Instruments, CHI660A, Austin, Texas). The electrodes were then immersed in 200 μmol L−1 ethanolic solutions of 11-mercaptoundecanoic acid, 11-mercaptoundecanol, 1-mercaptoundecane, or 11-mercaptoundecyl amine hydrochloride for 20 h in order to form a self-assembled monolayer on the gold surface. After rinsing with ethanol, a second CV of each electrode was recorded.Surface plasmon resonance (SPR) spectroscopy measurements
The SPR sensor chip consisting of a 50 nm Au layer on a 5 nm Cr adhesive layer on glass with a refractive index of 1.61 (Mivitec, Sinzing, Germany) was immersed in 200 μmol L−1 ethanolic solutions of 11-mercaptoundecanoic acid, 11-mercaptoundecanol, 1-mercaptoundecane, or 11-mercaptoundecyl amine hydrochloride for a minimum of 20 h in order to form a SAM on the gold surface. The chips were then rinsed with ethanol and dried under nitrogen flow. The SPR measurements were performed with a two channel SPR device (Mivitec Biosuplar 321, Sinzing, Germany) at ambient temperature with 640 nm laser excitation and equipped with a flow cell of a total volume of approx. 50 μL per channel. The flow rate was adjusted to 200 μL min−1. The measurements were performed at a fixed angle by read-out of the change in the intensity of the reflected light. After calibration of the measured signal intensity with sodium chloride solutions of known refractive index, liposome dispersions were diluted to phospholipid concentrations of 1, 5, 10, 25, 50, and 100 μmol L−1 with degassed HEPES-saline buffer. The dispersions were consecutively injected with increasing phospholipid concentration, each for 20 min followed by a washing step with HEPES-saline buffer for 10 min. The average signal changes and errors for the four different thiol surfaces were calculated from three independent measurements. For the measurement of blank gold, the error bars were obtained by taking 3× noise. For the temperature dependence studies the SPR device was placed in a thermally controlled room at 16 °C or a flow cell with temperature control was adjusted to 37 or 50 °C respectively. The calibration with the NaCl solutions was corrected with the adequate refractive index at these temperatures.Results and discussion
In this study, the binding interactions between two types of highly stable anionic liposomes with varying negative surface charge and differently modified gold surfaces are investigated via SPR spectroscopy (Fig. 1). Strong binding will result in high SPR signal changes due to the high refractive index of the lipid bilayer and liposome encapsulant (300 mM NaCl), whereas weak binding leads to only small changes.Liposomes were characterized with respect to their size and surface charge. Dynamic light scattering was used to determine their hydrodynamic diameters. The non-tagged liposomes had a diameter of 170 nm, with a polydispersity index (PDI) of 0.16. Liposomes with N-glutaryl-1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine (N-glutaryl-DPPE) are 240 nm in diameter (PDI of 0.22). The carboxylic groups providing additional negative surface charge as well as additional sodium counter ions can cause – among other synthesis parameters – such an increase in hydrodynamic diameter.35 Glutaryl-tags on the liposome surface lead to higher electrostatic repulsion between the liposomes. As a consequence, the incorporation of charged tags in the bilayer membrane contributes to improved colloidal stability. Furthermore, the higher surface charge can indeed enhance the colloidal stability in solution due to stronger repulsion between the nanovesicles, yet it can also increase binding to the surfaces of opposite surface charge, which needs to be considered when designing the overall sensor setup.
The zeta potential obtained by electrophoretic mobility measurements of –COOH-tagged liposomes (−47 ± 3 mV) is more negative than for the untagged ones (−34 ± 3 mV). Both types of liposomes show extreme long term stability and keep their physical and hydrodynamic diameters as well as the high negative zeta potential for at least 400 days.36 This exceeds by far the colloidal stability of numerous other nanomaterials such as polystyrene nanospheres37,38 or magnetite nanoparticles.39
Transmission electron microscopy (TEM) images show the successful formation of the liposomes that display good regularity of size and shape (Fig. 2). Deviations from the perfectly spherical shape of liposomes on the images are likely due to the drying and staining process on the TEM grid.
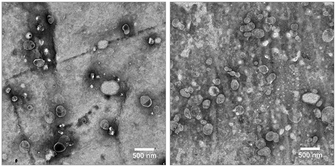 | ||
Fig. 2 TEM images of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted dispersions of untagged liposomes (left) and liposomes with the N-glutaryl-DPPE tag (right). Scale bars represent 500 nm. 10 diluted dispersions of untagged liposomes (left) and liposomes with the N-glutaryl-DPPE tag (right). Scale bars represent 500 nm. | ||
The phospholipid concentration was determined via ICP-AES resulting in 6.14 ± 0.12 mmol L−1 for the untagged batch and 11.17 ± 0.13 mmol L−1 for the liposomes with the N-glutaryl-DPPE tag. The total lipid concentration can be calculated using the molar ratios of the lipid composition (see the Experimental section) to be 11.3 ± 0.2 and 19.7 ± 0.2 mmol L−1, respectively. The total number of lipids per liposome is given by eqn (1):40
| Ntot = (π/aL)[d2 + (d − 2t)2] | (1) |
The successful formation of closely packed alkanethiol monolayers on gold provides an insulation barrier43 for the hexacyanoferrate and can be seen in a drastic change of the redox peaks in the cyclic voltammograms (CV) (Fig. S1†). Starting with clearly defined reduction peaks between +310 and +340 mV (vs. Ag/AgCl) and oxidation peaks between 0.05 and 0.12 mV for unmodified gold electrodes, all four different thiols show successful formation of a self-assembled monolayer (SAM) on the gold electrodes, as the surface is blocked for the redox reaction and the redox currents are significantly decreased, resulting in CVs without clearly discernible peak potentials. The best blocking is exhibited by the CH3-terminated thiol monolayer, followed by the –COOH- and –OH-modified surfaces. For the electrode modified with 11-mercaptoundecyl amine hydrochloride a hindered charge-transfer can be seen by the large separation of the peak potentials, indicating the formation of a monolayer.
Binding characteristics of anionic liposomes without –COOH tags
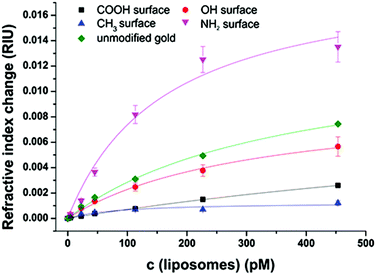
After extensive cleaning of the SPR gold chip and device calibration with sodium chloride solutions of known refractive index in order to make sure that the measured RI changes are accurate, the liposome dispersions in a concentration range of 0 to 100 μmol L−1 phospholipid content (corresponding to 0 to 453 pmol L−1 total liposome concentration and covering the concentration range usually used in analytical applications14,44) were allowed an interaction time of 20 min with the modified sensor surface in a continuous flow system, followed by a 10 min washing step to remove all loosely bound liposomes. The concentration dependent signal change in refractive index units (RIU) was compared for the blank gold surface and four self-assembled monolayers of long-chained alkanethiols with varying terminal groups of –CH3,–OH,–COOH, and –NH2 (Fig. 3).The binding curves were fitted to the extended Langmuir model following eqn (2):
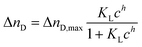 | (2) |
Positively cooperative binding is given by h > 1, and negatively cooperative binding by h < 1. For non-cooperative binding h is equal to 1. The fitting parameters are displayed in Tables S1 and S2† for both the simple Langmuir model (h fixed to 1), which neglects cooperative binding effects, and for the extended fit, which takes such effects into account. The Langmuir model was chosen over the Freundlich isotherm, as the experimental data display saturation behaviour.
The non-specific binding of anionic liposomes to varying surfaces can be mainly classified into two parameters: the total refractive index change at a certain liposome concentration and the binding affinity. Liposomes strongly bind to an unmodified gold surface resulting in a refractive index change of 0.007 RIU at 453 pM under physiological pH, as shown in Fig. 3. By modifying the gold surface with an amino-terminated monolayer, the binding interaction is even stronger and a signal change of 0.014 RIU can be observed. In the case of 11-mercaptoundecanol monolayers, signals (0.006 RIU) comparable to the unmodified gold were obtained. However, the 11-mercaptoundecanoic acid induces electrostatic repulsion leading to a clear blocking effect (0.003 RIU) which is even stronger for the hydrophobic surface (1-mercaptoundecane) which prevents the hydrophilic liposomes from binding (0.001 RIU).
Regarding the binding affinity, the standard Langmuir isotherms (h = 1) display decreasing dissociation constants with increasing positively charged sensor surface charge starting at KD = 1400 pM for HS(CH2)10–COO− due to the repulsive interactions. The uncharged hydrophilic surface self-assembled by HS(CH2)11–OH results in a lower KD value of 310 pM, whereas for the highly attractive interactions caused by the positively charged surface consisting of a SAM of HS(CH2)11–NH3+, a dissociation constant of 160 pM is obtained. For all hydrophilic surfaces, the refractive index change at maximum surface loading ΔnD, max lies consistently in the same range between 0.01 and 0.02 RIU, indicating a similar surface loading for the three surfaces at infinite liposome concentration. The SAM of the hydrophobic HS(CH2)10–CH3 on the other hand results in a very low ΔnD, max of 0.0013 RIU. This can be explained by the fact that the Langmuir fit is not very accurate for this hydrophobic surface (R2 = 0.85) due to the high differences in polarity between the liposome dispersions and the sensor surface. All other Langmuir fits show R2 values of more than 0.97 assuring the applicability of the model without the need to use more elaborate adsorption isotherms.
The standard Langmuir isotherms do not consider cooperative binding effects, especially for the interaction of positively charged amino groups to negatively charged phospholipids at pH 7.5. In accordance with this, the binding isotherms fitted by an extended Langmuir equation indicate positively cooperative binding for –NH2 SAM, non-cooperative binding in the case of –COOH SAM, negatively cooperative binding for blank gold and –OH SAM (Table S1†). The fitting of –CH3 SAM did not converge for this model, indicating that there is almost no interactions of the liposomes with this surface. In general, the coefficient referring to the cooperative effects is in all cases close to 1, and the precision of the binding constants is significantly lower than those obtained by the simple Langmuir isotherms.
Comparison of liposomes with and without the N-glutaryl-DPPE tag
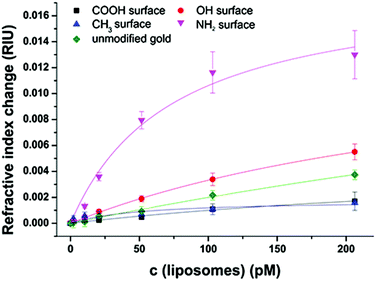
Liposomes offer a great variability in surface modification possibilities. As the starting point for conjugation to biomolecules N-glutaryl-DPPE is commonly used to add –COOH groups,45 with an even higher negative surface charge (zeta potential = −47 ± 3 mV) compared to liposomes without this tag (−34 ± 3 mV). The liposome dispersions in a concentration range of 0 to 100 μmol L−1 phospholipid content (corresponding to 0 to 206 pmol L−1 total liposome concentration) were allowed an interaction time of 20 min with the modified sensor surface in a continuous flow stream followed by a 10 min washing step to remove non-bound liposomes. The concentration dependent non-specific binding displayed by signal change in refractive index units (RIU) was again compared for the four model surfaces of –CH3, –OH, –COOH, and –NH2 and unmodified gold (Fig. 4). All binding curves are fitted to the simple and extended Langmuir model as before following eqn (2). The parameters resulting from the fit are listed in Table S2.†The very strong attractive interactions between the highly negatively charged liposomes with the cationic SAM consisting of HS(CH2)11–NH3+ lead to a high signal change of 0.012 RIU at a liposome concentration of 200 pmol L−1, whereas the uncharged hydroxyl surface results in 0.004 RIU (Fig. 4). Only very small changes can be observed for the interactions to the carboxyl- and the hydrophobic CH3-moiety (0.001 RIU). This is caused by the strong electrostatic repulsion of the first and the significant differences in polarity of the second surface.
As already observed for the untagged liposomes, the dissociation constants obtained from the Langmuir fits of the hydrophilic surfaces with the liposomes with the N-glutaryl-DPPE tag decrease from 400 pM for repulsive interaction of the carboxyl surface, over 330 pM for the 11-mercaptoundecanol to only 70 pM for the strong attractive interactions of the positively charged amino surface. The refractive index change at maximum surface loading lies in the range between 0.01 and 0.02 RIU for the hydroxyl- and amino-terminated surface and is significantly reduced to 0.0052 RIU for HS(CH2)10–COO− due to the strong electrostatic repulsion. The difference in polarity between the liposomes and HS(CH2)10–CH3 is the reason for the very low signal change at a maximum surface loading of 0.0018 RIU and the rather low R2 value of 0.72 for the fit. All other Langmuir fits show R2 values of more than 0.97 assuring the applicability of the model and showing no need of more elaborate fitting curves.
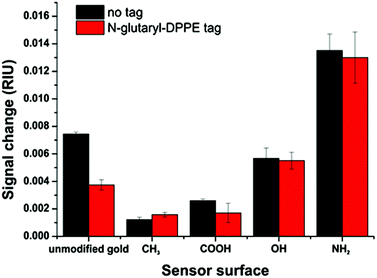
The additional negative surface charges introduced by the N-glutaryl-DPPE phospholipid influence the intra-molecular interactions between the charged liposomes themselves, thus reducing the apparent binding on the surfaces. Therefore, liposomes of the same phospholipid concentration of 100 μM were compared (translating into 453 pM liposome concentration for the untagged batch and 206 pM for the glutaryl-tagged batch), as this is a typical concentration used in analytical applications.14,45 For all surface modifications, except the –CH3 terminated surface, the signal change for the N-glutaryl-DPPE tagged liposomes is lower (Fig. 5). In the case of 11-mercaptoundecane all findings suggest that there is nearly no binding and therefore the signal changes are almost equal. However, the differences between the two types of liposomes are small in comparison with the influence of the modified sensor surface, showing the importance of careful sensor surface engineering for the prevention of non-specific adsorption of liposomes.
Additional –COOH tags provide the possibility of further functionalization and, due to the slightly more negative zeta potential, a better colloidal stability is achieved. In general, the lipid composition of the liposomes is already optimized for long term stability.36 The detailed study of the non-specific binding towards model surfaces, reflecting different chemical functionalities typically present in most biomolecules and sensor substrates, shows that the adhesion of this type of liposome is only weak. Even for positively charged amino surfaces the electrostatic repulsion between the liposomes themselves only leads to a low total surface coverage. Simulation of the binding of such liposomes neglecting their own electrostatic repulsion results in an expected shift in the SPR angle of at least 1.8°. This value is significantly higher than the shift found in the experiment for saturation (0.8°) (Fig. S2†). Liposomes consisting of only zwitterionic phospholipids, as primarily used in the studies by Kasemo et al., show significantly higher non-specific surface adsorption and tend to rupture to form a supported lipid bilayer.24,26 The low non-specific binding is therefore afforded by the overall lower binding ability and has to be taken into consideration in the design of analytical assays, i.e. the highly charged liposomes may ultimately be ideally suited for low analyte concentrations45 and microfluidic testing devices.15,29
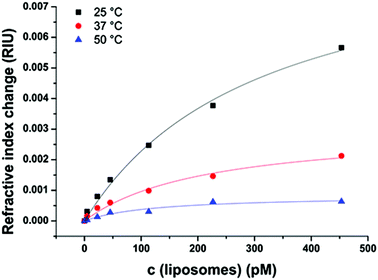
Temperature dependency of liposome binding
Binding events as well as the refractive index are strongly dependent on the temperature.46 Therefore, the interactions between the model surface consisting of HS(CH2)11–OH and liposomes without the N-glutaryl-DPPE tag were recorded at 25, 37 and 50 °C (Fig. 6), as these cover the temperature range at which most (bio)assays and (microfluidic) analytical applications with liposomes are carried out. The combination of untagged liposomes with the –OH terminated sensor surface was chosen to monitor the temperature dependence of the interactions unbiased by electrostatic interactions caused by the –NH2- and –COOH-modified surfaces. In addition, the difference between the liposomes with and without glutaryl-tag was the smallest when binding to the –OH surface. The same temperature-dependence studies were not duplicated with the tagged liposomes, because similar results can be expected in this small but relevant temperature range considering the almost identical values for KD and ΔnD, max (Tables S1 and S2†) for tagged and untagged liposomes when binding to the –OH surfaces. The surface with –CH3 terminal groups was not chosen due to the overall very low signal change upon liposome binding.The decrease in the overall signal change of liposomes binding to the sensor surface is in accordance with the decreasing refractive index of aqueous solutions with increasing temperature.46 The KD of the liposome binding determined by a simple Langmuir fit decreases between 25 and 50 °C from 310 ± 60 pM to 130 ± 50 pM, indicating stronger interactions between the liposomes and the model sensor surface with increasing membrane fluidity (Fig. S3†). Additionally, this displays the applicability of the Langmuir model over the whole examined range of temperatures.
Conclusions
The binding studies of anionic liposomes consisting of DPPC, DPPG and cholesterol with and without N-glutaryl-DPPE presented herein enable a better mechanistic understanding of the non-specific binding behaviour of these liposomes to model surfaces as used in all affinity-based (bio)analytical sensors or assays as well as point-of-care testing microfluidic devices. We showed that the binding events can be fitted to Langmuir isotherms with high R2 values (>0.97) for hydrophilic sensor surfaces. Thus, we were able to determine the KD values and the refractive index changes at maximum surface loading for these surface/liposome combinations.This is crucial when using liposomes as signal amplification tools,14,15,36,44 as non-specific binding can almost be prevented by modifying the sensor surface with –COOH groups. In addition, the introduction of 6 mol% N-glutaryl-DPPE into the mixed liposome surface reduces non-specific binding even further. This amount is large enough to provide electrostatic repulsion and high colloidal stability, but still small enough that specific binding is not prevented.44 This demonstrates that liposome surfaces can be tailored to a specific (analytical) task. They provide high specific binding on the one hand14 and show minimized non-specific binding to –COOH surfaces without the need of further protein blocking on the other hand, which makes them unique in comparison with other nanoparticle-based amplification systems, where such a surface engineering is by far not as straightforward. Furthermore, hydroxyl groups are known to minimize non-specific bindings, but we demonstrated that liposomes significantly adhere to –OH surfaces. This finding is important especially for miniaturized bioanalytical detection systems, such as microfluidic biosensors based on liposomes15,29 or similar nanoparticles with large surface areas compared to the total volume, where non-specific adsorption can cause significant problems and lead to high background signals and low sensing performance. Here, clever sensor surface engineering providing electrostatic repulsion or opposite polarity enables minimization of non-specific interactions and hopefully will lead, in the future, to intelligently blocked sensor surfaces and not bulk-blocking as customarily done through polymers and proteins. We will further study these conditions by investigations of the specific binding of receptor-modified liposomes to optimized surfaces. These findings herein will have a big impact on the general understanding of liposome and similar nanoparticle interactions and thus strongly contribute in the development of new and the improvement of existing applications using liposomes as drug carriers as well as for signal enhancement in sensing devices, ranging from standard microtiter plate assays to microfluidic point-of-care diagnostic tools.
Acknowledgements
We thank S. F. Himmelstoss for her graphical designs. We are grateful to J. Rewitzer und R. Walter for their technical assistance in the ICP-OES and SPR measurements. We thank R. Rachel for his expertise in TEM.References
- K. A. Edwards and A. J. Baeumner, Talanta, 2006, 68, 1421–1431 CrossRef CAS PubMed.
- F. Patolsky, A. Lichtenstein and I. Willner, J. Am. Chem. Soc., 2000, 122, 418–419 CrossRef CAS.
- M. Mukai, K. Maruo, Y. Sasaki and J. Kikuchi, Chem. – Eur. J., 2012, 18, 3258–3263 CrossRef PubMed.
- F. Patolsky, A. Lichtenstein and I. Willner, Angew. Chem., Int. Ed., 2000, 39, 940–943 CrossRef PubMed.
- Q. Liu and B. J. Boyd, Analyst, 2013, 138, 391–409 RSC.
- L. Zhang, F. Gu, J. Chan, A. Wang, R. Langer and O. Farokhzad, Clin. Pharmacol. Ther., 2008, 83, 761–769 CrossRef CAS PubMed.
- R. Tarallo, A. Accardo, A. Falanga, D. Guarnieri, G. Vitiello, P. Netti, G. D'Errico, G. Morelli and S. Galdiero, Chem. – Eur. J., 2011, 17, 12659–12668 CrossRef CAS PubMed.
- P. Pierrat, G. Laverny, G. Creusat, P. Wehrung, J.-M. Strub, A. VanDorsselaer, F. Pons, G. Zuber and L. Lebeau, Chem. – Eur. J., 2013, 19, 2344–2355 CrossRef CAS PubMed.
- B. Gruber and B. König, Chem. – Eur. J., 2013, 19, 438–448 CrossRef CAS PubMed.
- K. A. Edwards and A. J. Baeumner, Anal. Chem., 2014, 86, 6610–6616 CrossRef CAS PubMed.
- J. Zhou, Q. Wang and C. Zhang, J. Am. Chem. Soc., 2013, 135, 2056–2059 CrossRef CAS PubMed.
- S. Ahn and R. A. Durst, Anal. Bioanal. Chem., 2008, 391, 473–478 CrossRef CAS PubMed.
- S. Ahn-Yoon, T. R. DeCory and R. A. Durst, Anal. Bioanal. Chem., 2004, 378, 68–75 CrossRef CAS PubMed.
- C. Fenzl, T. Hirsch and A. J. Baeumner, Anal. Chem., 2015, 87, 11157–11163 CrossRef CAS PubMed.
- L. E. Locascio, J. S. Hong and M. Gaitan, Electrophoresis, 2002, 23, 799–804 CrossRef CAS PubMed.
- H. Liang, J.-P. Tuppurainen, J. Lehtinen, T. Viitala and M. Yliperttula, Eur. J. Pharm. Sci., 2013, 50, 492–501 CrossRef CAS PubMed.
- N. Granqvist, M. Yliperttula, S. Välimäki, P. Pulkkinen, H. Tenhu and T. Viitala, Langmuir, 2014, 30, 2799–2809 CrossRef CAS PubMed.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- C. F. Calver, H.-W. Liu and G. Cosa, Langmuir, 2015, 31, 11842–11850 CrossRef CAS PubMed.
- E. Tellechea, D. Johannsmann, N. F. Steinmetz, R. P. Richter and I. Reviakine, Langmuir, 2009, 25, 5177–5184 CrossRef CAS PubMed.
- I. Reviakine, M. Gallego, D. Johannsmann and E. Tellechea, J. Chem. Phys., 2012, 136, 84702 CrossRef PubMed.
- R. Richter, A. Mukhopadhyay and A. Brisson, Biophys. J., 2003, 85, 3035–3047 CrossRef CAS PubMed.
- R. P. Richter and A. R. Brisson, Biophys. J., 2005, 88, 3422–3433 CrossRef CAS PubMed.
- C. A. Keller and B. Kasemo, Biophys. J., 1998, 75, 1397–1402 CrossRef CAS PubMed.
- E. Reimhult, F. Höök and B. Kasemo, Langmuir, 2003, 19, 1681–1691 CrossRef CAS.
- E. Reimhult, M. Zäch, F. Höök and B. Kasemo, Langmuir, 2006, 22, 3313–3319 CrossRef CAS PubMed.
- P. He, L. Liu, W. Qiao and S. Zhang, Chem. Commun., 2014, 50, 1481–1484 RSC.
- C.-H. Yoon, J.-H. Cho, H.-I. Oh, M.-J. Kim, C.-W. Lee, J.-W. Choi and S.-H. Paek, Biosens. Bioelectron., 2003, 19, 289–296 CrossRef CAS PubMed.
- N. Wongkaew, P. He, V. Kurth, W. Surareungchai and A. J. Baeumner, Anal. Bioanal. Chem., 2013, 405, 5965–5974 CrossRef CAS PubMed.
- K. Dimitrievski and B. Kasemo, Langmuir, 2009, 25, 8865–8869 CrossRef CAS PubMed.
- V. Shpacovitch, V. Temchura, M. Matrosovich, J. Hamacher, J. Skolnik, P. Libuschewski, D. Siedhoff, F. Weichert, P. Marwedel, H. Müller, K. Überla, R. Hergenröder and A. Zybin, Anal. Biochem., 2015, 486, 62–69 CrossRef CAS PubMed.
- K. A. Edwards and A. J. Baeumner, Anal. Bioanal. Chem., 2006, 386, 1335–1343 CrossRef CAS PubMed.
- V. Bello, G. Mattei, P. Mazzoldi, N. Vivenza, P. Gasco, J. M. Idee, C. Robic and E. Borsella, Microsc. Microanal., 2010, 16, 456–461 CrossRef CAS PubMed.
- R. J. Hunter, Zeta Potential in Colloid Science: Principles and Applications, Academic Press, London, 1981 Search PubMed.
- J. Sabın, G. Prieto, J. M. Ruso, R. Hidalgo-Álvarez and F. Sarmiento, Eur. Phys. J. E, 2006, 20, 401–408 CrossRef PubMed.
- K. A. Edwards, K. J. Meyers, B. Leonard and A. J. Baeumner, Anal. Bioanal. Chem., 2013, 405, 4017–4026 CrossRef CAS PubMed.
- C. Fenzl, S. Wilhelm, T. Hirsch and O. S. Wolfbeis, ACS Appl. Mater. Interfaces, 2012, 5, 173–178 Search PubMed.
- C. Fenzl, C. Genslein, A. Zöpfl, A. J. Baeumner and T. Hirsch, J. Mater. Chem. B, 2015, 3, 2089–2095 RSC.
- P. Fraga García, M. Freiherr von Roman, S. Reinlein, M. Wolf and S. Berensmeier, ACS Appl. Mater. Interfaces, 2014, 6, 13607–13616 Search PubMed.
- A. K. Singh, P. K. Kilpatrick and R. G. Carbonell, Biotechnol. Prog., 1996, 12, 272–280 CrossRef CAS PubMed.
- C. Ege and K. Y. C. Lee, Biophys. J., 2004, 87, 1732–1740 CrossRef CAS PubMed.
- J. N. Israelachvili and D. J. Mitchell, Biochim. Biophys. Acta, Biomembr., 1975, 389, 13–19 CrossRef CAS.
- P. Diao, D. Jiang, X. Cui, D. Gu, R. Tong and B. Zhong, J. Electroanal. Chem., 1999, 464, 61–67 CrossRef CAS.
- K. A. Edwards and A. J. Baeumner, Anal. Chem., 2013, 85, 2770–2778 CrossRef CAS PubMed.
- K. A. Edwards, K. J. Meyers, B. Leonard, J. T. Connelly, Y. Wang, T. Holter and A. J. Baeumner, Anal. Methods, 2012, 4, 3948–3955 RSC.
- P. N. Yi and R. C. MacDonald, Chem. Phys. Lipids, 1973, 11, 114–134 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6an00820h |
| This journal is © The Royal Society of Chemistry 2016 |