Open Access Article
Open Access ArticleEmerging techniques in the isolation and characterization of extracellular vesicles and their roles in cancer diagnostics and prognostics
Vijaya
Sunkara†
a,
Hyun-Kyung
Woo†
a and
Yoon-Kyoung
Cho
*ba
aDepartment of Biomedical Engineering, School of Life Sciences, Ulsan National Institute of Science and Technology (UNIST), UNIST-gil 50, Ulsan 689-798, Republic of Korea. E-mail: ykcho@unist.ac.kr
bCenter for Soft and Living Matter, Institute for Basic Science (IBS), UNIST-gil 50, Ulsan 689-798, Republic of Korea. E-mail: ykcho@unist.ac.kr
First published on 26th October 2015
Abstract
Extracellular vesicles (EVs) are cell-derived nanovesicles, present in almost all types of body fluids, which play an important role in intercellular communication and are involved in the transport of biological signals for regulating diverse cellular functions. Due to the increasing clinical interest in the role of EVs in tumor promotion, various techniques for their isolation, detection, and characterization are being developed. In this review, we present an overview of the current EV isolation and characterization methods in addition to their applications and limitations. Furthermore, EVs as the potential emerging biomarkers in cancer management and their clinical implementation are briefly discussed.
1. Introduction
Cells release several types of extracellular vesicles (EVs) including exosomes (40–120 nm in diameter, released during the fusion of multi-vesicular endosomes with the plasma membrane), microvesicles (100–1000 nm in diameter, formed by direct budding from the plasma membrane) and apoptotic bodies (1–5 μm in diameter, produced by cells due to apoptosis), which have been classified based on their biogenesis and size.1–4 However, no analysis method or specific marker is currently available to distinguish the origin of these vesicles, upon release into the extracellular matrix. Furthermore, it is difficult to differentiate between vesicles of a similar size when they have been isolated together. Different terminologies such as EVs, microvesicles, or exosomes are used synonymously, particularly in the engineering community, for the isolated nanovesicles and there is no consensus available to discriminate these vesicles. Therefore, in this review, we will use the term “EVs” for all types of vesicles that measure <200 nm in diameter, to make the review simple and understandable.EVs are secreted by most cell types and are present in many kinds of body fluids such as blood plasma, urine, amniotic fluid, saliva, ascites, cerebrospinal fluid etc.5 At the time of their discovery, they were thought to be involved in cellular excretion of by-products and not much attention was given to them until their participation in the immune response was revealed.6,7 In 2007, it was shown that they mediate communication between cells due to the presence of functional RNAs.8 After that, research on EVs has grown exponentially focusing on the development of new techniques for the isolation as well as analysis of their composition and physiological functions.4,9,10
The functions of EVs depend on their origin as well as their composition. Because they carry diverse membrane and cytosolic proteins, DNA, mRNA and miRNA, EVs are considered potential biomarkers and are found to be involved in both normal physiological processes and disease progression in a pleiotropic manner, including the immune response, antigen presentation, cell signaling, intercellular communication etc.2,9,11–20
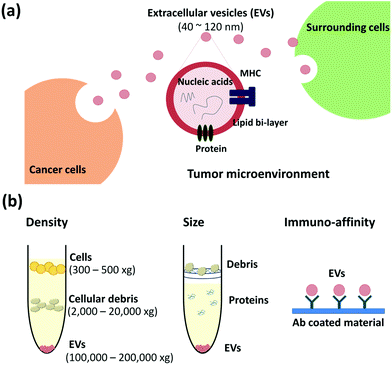
In a tumor microenvironment, communication between cancer and surrounding cells is essential for the tumors to survive, metastasize and spread at distant sites (Fig. 1a). Here, EVs play a vital role to transfer the tumor related genetic information to the surrounding as well as distant cells and promote tumor progress through metastatic niche formation, tumor stroma generation and angiogenesis.9,11,20–24
In cancer research, intercellular communication through miRNA is of particular interest, as they can regulate the gene expression in recipient cells.25–32 However, the amount of miRNA from EVs and its clinical importance remain a matter of debate.23,33 In the literature, it is still not clear which miRNA, the free miRNA or the exosomal miRNA, contributes to a greater extent. Nonetheless, the advantage of isolating and analyzing the exosomal RNAs is that the lipid bilayer protects these molecules from degradation and allows efficient recovery from biological fluids.34,35
Isolation and analysis of pure EVs further enhance our understanding of their roles in numerous biological events. As EVs are gaining importance as biomarkers of tumor signatures, their isolation with high purity is necessary, or else their analysis could yield misleading results owing to the presence of contaminants such as viruses, lipoproteins, large protein aggregates, and other vesicles. Differential ultracentrifugation is the conventional and most widely used method for the isolation of different vesicles from bio-fluids. However, it requires a long time for isolation and different particles with similar physical properties can co-sediment. Thus, to overcome these limitations, sucrose gradient purification and precipitation reagents were developed. Numerous groups have been working on developing microfluidic devices for the efficient isolation of pure EVs from bio-fluids.10
The size of EVs is much below the resolution of conventional fluorescence microscopes. Therefore, specialized equipment is needed for the detection and analysis of the physical characteristics of the EVs. At the same time, characterizing the exosomal content is very important for understanding biological functions of EVs as well as their roles in cancer development. Several techniques are available and a combination of different methods is now being used for specific EV analysis.36 Recent reviews discuss EVs as an emerging biomarker for intercellular communication and medical diagnostics.4,12–15 In addition, the advances in isolation technologies of exosomes using micro and nanofluidic devices were reviewed recently.10,37 However, Liga et al. mainly focused on various isolation techniques whereas Ko et al. reviewed the isolation as well as detection methods, but the surface marker and protein analysis was not discussed in detail. Here, we will focus on recent trends in EV isolation and detection along with the surface characterization methods and their roles in cancer diagnostics and prognostics.
2. EV isolation methods
With the growing interest in EVs as potential biomarkers, various isolation techniques have recently been developed.10 Furthermore, several research groups compared the efficiency of available isolation techniques.38–40 Based on the principle of their separation, the methods can be categorized into three major classes: density-, size-, and immunoaffinity-based isolation. Here, we have summarized the common isolation techniques and recent trends along with new approaches.2.1 Density-based isolation
As the name suggests, based on their density, particles are separated upon centrifugation. The techniques include ultracentrifugation and regular centrifugation with precipitation reagents including commercialized kits (Fig. 1b, left). These techniques enable the concentration of EVs rather than isolation.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g sediments the EVs (1.13–1.19 g mL−1).11,41 UC is a conventional and most commonly used method for isolating EVs. However, it takes ∼5 h for isolating the exosomal pellet;5,42 moreover, it requires special equipment, an ultracentrifuge, which might be unavailable in common laboratories in clinical settings. Other drawbacks include inefficient isolation and low recovery.38–40 Therefore, sucrose gradient centrifugation has been developed to improve the purity and recovery.38 Nevertheless, it cannot separate impurities like viruses43 and also it is time-consuming owing to complicated sample-processing steps with differential centrifugation.
000g sediments the EVs (1.13–1.19 g mL−1).11,41 UC is a conventional and most commonly used method for isolating EVs. However, it takes ∼5 h for isolating the exosomal pellet;5,42 moreover, it requires special equipment, an ultracentrifuge, which might be unavailable in common laboratories in clinical settings. Other drawbacks include inefficient isolation and low recovery.38–40 Therefore, sucrose gradient centrifugation has been developed to improve the purity and recovery.38 Nevertheless, it cannot separate impurities like viruses43 and also it is time-consuming owing to complicated sample-processing steps with differential centrifugation.
2.2 Size-based isolation
With exosomal size being known (<200 nm), EVs can be separated from cells and large debris by using nano-sized membrane filters (Fig. 1b, middle). Filtration is an effective method for the removal of large debris including large vesicles (above 200 nm in diameter), sub-cellular fractions, protein aggregates, protein–nucleic acid aggregates, or plasma proteins.47 Membranes with pore sizes of 0.2, 0.22, and 0.45 μm and materials such as polyvinylidene fluoride48 and track-etched polycarbonate are widely used to remove large debris.49,50 The size based isolation techniques also enrich the EVs from a complex mixture of samples but do not isolate them specifically.To enhance the purity, a combination of filtration and UC is also commonly used.23,51–53 Muller et al. reported that this combined method is better than using a UC based method, differential centrifugation or the sucrose density gradient method alone.47 Davies et al. applied the combined method with pressure- and electrophoresis driven filtration for isolating and purifying EVs.54
2.3 Affinity-based isolation
To isolate EVs and study their relationship with cancer, new isolation techniques utilizing microfluidics including paper-based microfluidic devices, immunomagnetic beads, photosensitizer beads etc. are being developed, based on immunoaffinity capture with antibodies specific to EVs (Fig. 1b, right).38,49,61–64 The common exosomal markers are tetraspanins including CD9, CD63, and CD81 and tumor-associated markers are epithelial cell adhesion molecules (EpCAM), IGF-1R α units (α-IGF-1R), CA125, CD41b, and E-cadherin.42,65 With the markers specific to exosomes, these methods aid in the specific isolation of exosomes depending on the capture antibodies used.Various microfluidic devices are developed for selective isolation and analysis on a single device by using a small sample volume and within a short period of time. Due to their simplicity, these techniques can be applied to point of care (POC) diagnostics. The microfluidic chip shown in Fig. 2a uses magnetic microbeads (3 μm in diameter) coated with anti-EGFR to capture EVs from 100 μL of serum and has >93% capture efficiency. It is an integrated method from EV isolation to RNA analysis by qPCR in a single chip, which can be completed within 2 h.58 Electric field-induced release and measurement (EFIRM) was used for disrupting exosomes and monitoring the released RNA/protein biomarkers, after capturing the EVs by using anti-CD63-magnetic beads (Fig. 2b). With this device and in vivo experiments, the authors confirmed that the H460-CD63-GFP shed exosomes were transported to blood and saliva.59 In another approach, He et al. used magnetic beads coated with anti-EpCAM and anti-CA125 to isolate EVs in volumes as low as 30 μL of plasma. It is an integrated analysis technique including exosome capture and protein detection using chemi-fluorescence in a single chip.65 Yoshioka et al. used photosensitizer-beads to capture CD147- and CD9-positive EVs from 5 μL of serum, as shown in Fig. 2c. They could complete EV capture and analysis within 1.5 h by using ExoScreen.60 Zhu et al. used an antibody microarray with surface plasmon resonance imaging (SPRi) to capture and detect tumor-derived EVs in real time and in a label-free manner, as shown in Fig. 2d.42
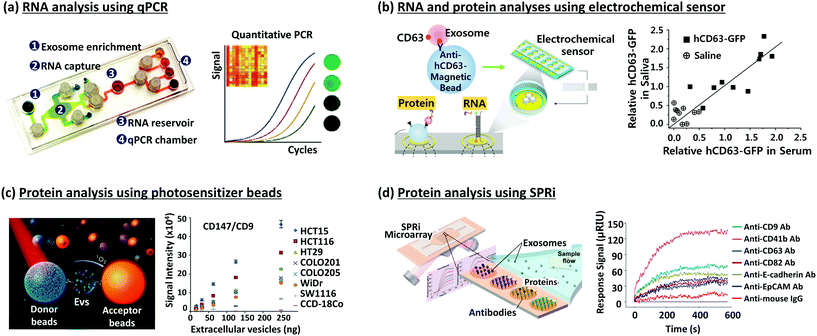 | ||
| Fig. 2 Devices for EV isolation and analysis. (a) RNA analysis using qPCR.58 (b) RNA and protein analysis using an electrochemical sensor.59 (c) Protein analysis using photosensitizer beads.60 (d) Protein analysis using SPRi.42 Reproduced from ref. 42 and 58–60 with permission from the Nature Publishing Group, Elsevier, and American Chemical Society. | ||
Apart from the above techniques, the methods used for the separation of nanoparticles have a high potential to be used in the isolation of EVs from a complex mixture of samples. For example, the Heller group developed a dielectrophoretic method for the recovery of drug delivery nanoparticles as well as cancer related biomarkers from plasma and blood.66,67 This approach can be used for the isolation and detection of EVs as well.
3. Detection and downstream analysis of EVs
Novel detection methods are required to explore the physical characteristics and biology of EVs. To analyze them, a combination of different optical and non-optical techniques is utilized, including electron microscopy, light scattering, fluorescence, and molecular profiling. Here, we have summarized the most commonly used methods, their applicability, and limitations.3.1 Size characterization
Size and morphological information can be obtained from high-resolution imaging with electron microscopy utilizing transmission electron microscopy (TEM), scanning electron microscopy (SEM),70 or atomic force microscopy (AFM) (Fig. 3a). The light scattering techniques (Fig. 3b–d) determine the relative size distribution in a solution, the concentration, and the zeta potential of the particles.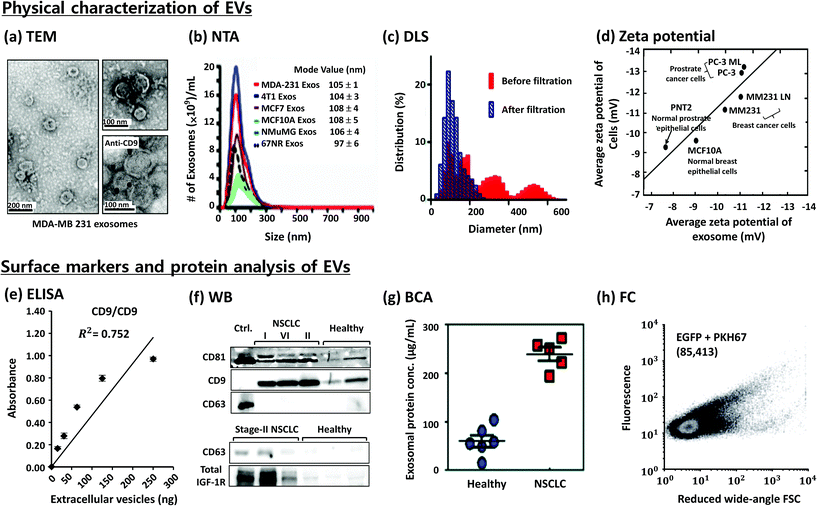 | ||
| Fig. 3 Characterization of EVs. (a) Evaluation of morphology by transmission electron microscopy (TEM)23 and (b) size distribution and concentration by nanoparticle tracking analysis (NTA),23 (c) size distribution by dynamic light scattering (DLS),49 (d) zeta potential measurements,68 (e) enzyme-linked immunosorbent assay (ELISA),60 (f) western blotting (WB),65 (g) Bradford assay for total protein quantification (BCA) of EVs from non-small-cell lung cancer (NSCLC) patients and healthy people,65 and (h) flow cytometry (FC).69 Reproduced from ref. 23, 49, 60, 65, 68, and 69 with permission from Elsevier, Nature Publishing Group, Royal Society of Chemistry and Japan Society of Applied Physics. | ||
Zeta potential indicates the relative stability of the particles in solution.77 It is determined by measuring their velocity during the electrophoresis. Charged particles migrate towards the electrode under an applied electric field. The speed of the particles is proportional to the field strength and the zeta potential. The value of the zeta potential depends on the temperature, pH, conductivity, and salt concentration of the buffer used.
DLS analysis gives the average size of relatively mono-disperse populations of isolated EVs, and it may not be a good choice for a heterogeneous mixture of EVs. No difference was observed between the average sizes of EVs derived from cancer cells compared to those of EVs derived from normal cells. However, the EVs derived from cancer cells showed higher zeta potential than those derived from normal cells when measured using an on-chip micro capillary electrophoresis system (Fig. 3d).68
3.2 Surface markers and protein analysis
Several methods including ELISA, western blotting, Bradford's assay, and flow cytometry provide information on the presence of proteins and specific surface markers (Fig. 3e–h). However, standardization and optimization of the protocols are necessary before being adapted for the analysis.3.3 Molecular analysis
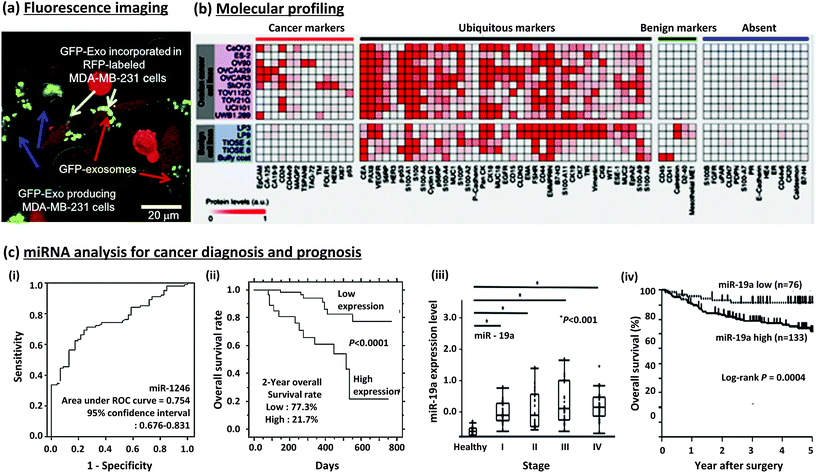 | ||
| Fig. 4 EVs in a tumor microenvironment and their protein/miRNA analysis for cancer diagnosis and prognosis. (a) Fluorescence images of cells producing GFP-labelled EVs and their uptake by recipient cells.75 (b) Molecular profiling of protein markers.49 (c) (i & ii) Detection and prognosis of miR1246 in esophageal squamous cell carcinoma (ESCC).76 (iii & iv) Detection and prognosis of miR-19a expression in colorectal cancer (CRC).32 Reproduced from ref. 32, 49, 75 and 76 with permission from Elsevier and the Nature Publishing Group. | ||
Quantitative reverse transcription polymerase chain reaction (qRT PCR), miRNA arrays, bioanalyzers and mass spectrometry are widely used for profiling exosomal cargo.45,97,98 Wyss et al. used single fluctuation analysis to profile both the size and the relative expression level of a particular membrane receptor in individual EV.87 Rekker et al. compared isolation methods for microRNA profiling, showing that both UC and ExoQuick precipitation methods are suitable for profiling.45
Im et al. conducted molecular profiling of ovarian cancer protein markers, as shown in Fig. 4b. Through this, ovarian cancer cells and benign cells could be differentiated from each other.49 Ueda et al. established mass spectrometric immunoassay monolith pipette tips to profile serum EV, identifying CD91 as a lung adenocarcinoma specific antigen on EV.98
| Cancer types | Sample type | EV isolation method | RNA isolation | Exosomal contents (miRNA) | Clinical value |
|---|---|---|---|---|---|
| Esophageal squamous cell carcinoma (ESCC) | Serum | Ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
TRIzol reagent, mirVana PARIS kit | miR-1246 | Upregulation in patients76 |
| Serum | 0.45 μm filtration, ExoQuick | mirVana microRNA isolation kit (Life Tech) | miR-21 | Upregulation in patients than benign disease48 | |
| Colorectal cancer (CRC) | Serum | Ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
Acid-guanidine-phenol-chloroform | miR-19a and 92a | Upregulation in patients. Higher expression is indicative of poor prognosis32 |
| Serum | 0.2 μm filtration, ultracentrifugation (120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
TRIzol reagent (Invitrogen), RNeasy Mini spin column (Qiagen), Agilent miRNA labeling reagent | Let-7a, miR-1229, 1246, 150, 21, 223, 23a | Upregulation in patients26 | |
| Hepato-cellular carcinoma (HCC) | Serum | 0.22 μm filtration, ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
miRNeasy Mini Kit (Qiagen) | miR-718 (tumor-suppressive miRNA) | Progression and recurrence. |
| Downregulation in patients with recurrence compared to that observed in those without recurrence78 | |||||
| Serum | Total exosome isolation reagent (Invitrogen) | Isothiocyanate-phenol/chloroform extraction procedures | miR-21 | Upregulation in patients compared to that observed in patients with hepatitis or in healthy controls79 | |
| Breast cancer | Serum | ExoQuick exosome precipitation solution (BioCat) | mirVana PARIS kit (Life Tech) | miR-101, 372, 373 | Upregulation in patients80 |
| Cervical cancer | Cervicovaginal lavages | Ultracentrifugation | mirVana microRNA isolation kit (Ambion) | miR-21, 146a | Upregulation in patients29 |
| Pancreatic cancer | Serum | Sucrose-gradient centrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
miRNeasy Mini Kit (Qiagen) | miR-1246, 4644, 3976, 4306 | Upregulation in patients81 |
| Prostate cancer | Plasma | ExoQuick exosome precipitation solution (SBI) | miRNeasy Micro Kit (Qiagen) | miR-1290, miR-375 | Upregulation in patients. Their levels are associated with a poor survival rate82 |
| Cancer types | Sample type | EV isolation method | Protein assay | Exosomal contents (protein) | Clinical value |
|---|---|---|---|---|---|
| Pancreatic cancer | Serum | 0.2 μm filtration, ultracentrifugation (150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
Ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) | Glypican-1 (GPC1) | 100% sensitivity and specificity for detecting pancreatic cancer92 |
| Saliva | Magnetic beads based capture | Western blotting | Apbblip, Daf2, Foxp1, Incenp, BCO31781, Aspn, and Gng2 | Upregulation in pancreatic cancer-derived exosomes from tumor-bearing mice93 | |
| Acute myeloid leukemia (AML) | Plasma | 0.22 μm filtration, Bio-Gel A50 m columns (Bio-Rad), ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
Immunoaffinity capture | CD34 | Upregulation in patients94 |
| Colon cancer | Serum | Immunoaffinity capture | ExoScreen | CD147, CD9 | Upregulation of double positive (both CD147 and CD9) in patients60 |
| Prostate cancer | Plasma | Ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
Western blotting | Phosphatase and tensin homolog (PTEN) | Only detected in cancer patients95 |
| Bladder cancer | Urine | 0.2 μm filtration, ultracentrifugation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) 000g) |
Western blotting | EDIL-3/Del1 | Upregulation in patients associated with tumor grade96 |
4. Conclusions and perspectives
EVs contain a substantial amount of genetic information that could be transferred to other cells, thereby promoting the progression of metastatic tumors in cancer patients. Several methods are being developed to isolate and analyze EVs to understand their physiological functions. However, still there is a need for new technologies that could isolate pure EVs from a mixture of various particles and contaminants.One of the biggest issues in the EV research is the lack of proper terminology and methodology. Though we termed all vesicles as EVs in this article, there should be a clear demarcation for each vesicle and the data regarding their content, specific markers and physiological functions should be obtained and compared for different kinds of vesicles.
Another major issue in this field is differentiating between microvesicles and exosomes of similar size. Any vesicles that share size, density, or scattering properties would interfere with the isolation and detection of other vesicles. Devices that specifically capture a particular type of vesicles should be developed.
In addition, efficiency in terms of purification and quantification of the currently available isolation procedures is unknown. A standard must be developed to compare the data of different sets of experiments.
Apart from all these issues, demonstrating the physiological functions of EVs in vivo is a major challenge. Thus far, data regarding EVs have been obtained from in vitro studies and very little is known about their function in vivo. Nevertheless, increasing evidence suggests that EVs are emerging biomarkers for cancer diagnosis and therapeutics.
Acknowledgements
This work was supported by the National Research Foundation (NRF) grants (2013R1A2A2A05004314, 2012R1A1A2043747), a grant from the Korean Health Technology R&D Project, the Ministry of Health & Welfare (A121994) and IBS-R020-D1 funded by the Korean Government.References
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS PubMed.
- V. R. Minciacchi, M. R. Freeman and D. Di Vizio, Semin. Cell Dev. Biol., 2015, 40, 41–51 CrossRef CAS PubMed.
- J. Kowal, M. Tkach and C. Théry, Curr. Opin. Cell Biol., 2014, 29, 116–125 CrossRef CAS PubMed.
- S. E. Andaloussi, I. Mäger, X. O. Breakefield and M. J. Wood, Nat. Rev. Drug Discovery, 2013, 12, 347–357 CrossRef PubMed.
- K. W. Witwer, E. I. Buzás, L. T. Bemis, A. Bora, C. Lässer, J. Lötvall, E. N. Nolte-'t Hoen, M. G. Piper, S. Sivaraman, J. Skog, C. Théry, M. H. Wauben and F. Hochberg, J. Extracell. Vesicles, 2013, 2, 20360 Search PubMed.
- G. Raposo, H. W. Nijman, W. Stoorvogel, R. Liejendekker, C. V. Harding, C. J. Melief and H. J. Geuze, J. Exp. Med., 1996, 183, 1161–1172 CrossRef CAS PubMed.
- L. Zitvogel, A. Regnault, A. Lozier, J. Wolfers, C. Flament, D. Tenza, P. Ricciardi-Castagnoli, G. Raposo and S. Amigorena, Nat. Med., 1998, 4, 594–600 CrossRef CAS PubMed.
- H. Valadi, K. Ekström, A. Bossios, M. Sjöstrand, J. J. Lee and J. O. Lötvall, Nat. Cell Biol., 2007, 9, 654–659 CrossRef CAS PubMed.
- L. Brinton, H. Sloane, M. Kester and K. Kelly, Cell. Mol. Life Sci., 2015, 72, 659–671 CrossRef CAS PubMed.
- A. Liga, A. D. B. Vliegenthart, W. Oosthuyzen, J. W. Dear and M. Kersaudy-Kerhoas, Lab Chip, 2015, 15, 2388–2394 RSC.
- X. Zhang, X. Yuan, H. Shi, L. Wu, H. Qian and W. Xu, J. Hematol. Oncol., 2015, 8, 1–13 CrossRef CAS PubMed.
- E. I. Buzas, B. Gyorgy, G. Nagy, A. Falus and S. Gay, Nat. Rev. Rheumatol., 2014, 10, 356–364 CrossRef CAS PubMed.
- C. Soekmadji, P. Russell and C. Nelson, Cancers, 2013, 5, 1522–1544 CrossRef CAS PubMed.
- A.-K. Ludwig and B. Giebel, Int. J. Biochem. Cell Biol., 2012, 44, 11–15 CrossRef CAS PubMed.
- P. Kharaziha, S. Ceder, Q. Li and T. Panaretakis, Biochim. Biophys. Acta, Rev. Cancer, 2012, 1826, 103–111 CrossRef CAS PubMed.
- T. Katsuda, N. Kosaka and T. Ochiya, Proteomics, 2014, 14, 412–425 CrossRef CAS PubMed.
- J. Rak and A. Guha, BioEssays, 2012, 34, 489–497 CrossRef CAS PubMed.
- A. V. Vlassov, S. Magdaleno, R. Setterquist and R. Conrad, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 940–948 CrossRef CAS PubMed.
- A. L. S. Revenfeld, R. Bæk, M. H. Nielsen, A. Stensballe, K. Varming and M. Jørgensen, Clin. Ther., 2014, 36, 830–846 CrossRef PubMed.
- Y. Sun and J. Liu, Clin. Ther., 2014, 36, 863–872 CrossRef PubMed.
- Y. Zhang and X.-F. Wang, Nat. Cell Biol., 2015, 17, 709–711 CrossRef CAS PubMed.
- B. Costa-Silva, N. M. Aiello, A. J. Ocean, S. Singh, H. Zhang, B. K. Thakur, A. Becker, A. Hoshino, M. T. Mark and H. Molina, Nat. Cell Biol., 2015, 17, 816–826 CrossRef CAS PubMed.
- S. A. Melo, H. Sugimoto, J. T. O'Connell, N. Kato, A. Villanueva, A. Vidal, L. Qiu, E. Vitkin, L. T. Perelman and C. A. Melo, Cancer Cell, 2014, 26, 707–721 CrossRef CAS PubMed.
- A. Azmi, B. Bao and F. Sarkar, Cancer Metastasis Rev., 2013, 32, 623–642 CrossRef CAS PubMed.
- M. Ono, N. Kosaka, N. Tominaga, Y. Yoshioka, F. Takeshita, R.-u. Takahashi, M. Yoshida, H. Tsuda, K. Tamura and T. Ochiya, Sci. Signaling, 2014, 7, 63 CrossRef PubMed.
- H. Ogata-Kawata, M. Izumiya, D. Kurioka, Y. Honma, Y. Yamada, K. Furuta, T. Gunji, H. Ohta, H. Okamoto and H. Sonoda, PLoS One, 2014, 9, e92921 Search PubMed.
- S. Rana, K. Malinowska and M. Zöller, Neoplasia, 2013, 15, 281–295 CrossRef CAS PubMed.
- D. M. Cereghetti and P. P. Lee, Microrna, 2013, 2, 194–204 CrossRef CAS.
- J. Liu, H. Sun, X. Wang, Q. Yu, S. Li, X. Yu and W. Gong, Int. J. Mol. Sci., 2014, 15, 758–773 CrossRef PubMed.
- B. N. Hannafon and W.-Q. Ding, Int. J. Mol. Sci., 2013, 14, 14240–14269 CrossRef PubMed.
- I. Hoshino and H. Matsubara, Surg. Today, 2013, 43, 467–478 CrossRef CAS PubMed.
- T. Matsumura, K. Sugimachi, H. Iinuma, Y. Takahashi, J. Kurashige, G. Sawada, M. Ueda, R. Uchi, H. Ueo and Y. Takano, Br. J. Cancer, 2015, 115, 275–281 CrossRef PubMed.
- J. R. Chevillet, Q. Kang, I. K. Ruf, H. A. Briggs, L. N. Vojtech, S. M. Hughes, H. H. Cheng, J. D. Arroyo, E. K. Meredith, E. N. Gallichotte, E. L. Pogosova-Agadjanyan, C. Morrissey, D. L. Stirewalt, F. Hladik, E. Y. Yu, C. S. Higano and M. Tewari, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14888–14893 CrossRef CAS PubMed.
- Q. Li, Y. Shao, X. Zhang, T. Zheng, M. Miao, L. Qin, B. Wang, G. Ye, B. Xiao and J. Guo, Tumor Biol., 2015, 36, 2007–2012 CrossRef CAS PubMed.
- Q. Ge, Y. Zhou, J. Lu, Y. Bai, X. Xie and Z. Lu, Molecules, 2014, 19, 1568–1575 CrossRef PubMed.
- E. van der Pol, F. Coumans, Z. Varga, M. Krumrey and R. Nieuwland, J. Thromb. Haemostasis, 2013, 11, 36–45 CrossRef PubMed.
- J. Ko, E. Carpenter and D. Issadore, Analyst, 2015 10.1039/C5AN01610J.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Methods, 2012, 56, 293–304 CrossRef CAS PubMed.
- J. Caradec, G. Kharmate, E. Hosseini-Beheshti, H. Adomat, M. Gleave and E. Guns, Clin. Biochem., 2014, 47, 1286–1292 CrossRef CAS PubMed.
- R. E. Lane, D. Korbie, W. Anderson, R. Vaidyanathan and M. Trau, Sci. Rep., 2015, 5, 7639 CrossRef PubMed.
- W. Oosthuyzen, N. E. Sime, J. R. Ivy, E. J. Turtle, J. M. Street, J. Pound, L. E. Bath, D. J. Webb, C. D. Gregory and M. A. Bailey, J. Physiol., 2013, 591, 5833–5842 CrossRef CAS PubMed.
- L. Zhu, K. Wang, J. Cui, H. Liu, X. Bu, H. Ma, W. Wang, H. Gong, C. Lausted and L. Hood, Anal. Chem., 2014, 86, 8857–8864 CrossRef CAS PubMed.
- R. Cantin, J. Diou, D. Bélanger, A. M. Tremblay and C. Gilbert, J. Immunol. Methods, 2008, 338, 21–30 CrossRef CAS PubMed.
- D. H. Atha and K. C. Ingham, J. Biol. Chem., 1981, 256, 12108–12117 CAS.
- K. Rekker, M. Saare, A. M. Roost, A.-L. Kubo, N. Zarovni, A. Chiesi, A. Salumets and M. Peters, Clin. Biochem., 2014, 47, 135–138 CrossRef CAS PubMed.
- H. Shin, C. Han, J. M. Labuz, J. Kim, J. Kim, S. Cho, Y. S. Gho, S. Takayama and J. Park, Sci. Rep., 2015, 5, 13103 CrossRef CAS PubMed.
- L. Muller, C.-S. Hong, D. B. Stolz, S. C. Watkins and T. L. Whiteside, J. Immunol. Methods, 2014, 411, 55–65 CrossRef CAS PubMed.
- Y. Tanaka, H. Kamohara, K. Kinoshita, J. Kurashige, T. Ishimoto, M. Iwatsuki, M. Watanabe and H. Baba, Cancer, 2013, 119, 1159–1167 CrossRef CAS PubMed.
- H. Im, H. Shao, Y. I. Park, V. M. Peterson, C. M. Castro, R. Weissleder and H. Lee, Nat. Biotechnol., 2014, 32, 490–495 CrossRef CAS PubMed.
- J. Rho, J. Chung, H. Im, M. Liong, H. Shao, C. M. Castro, R. Weissleder and H. Lee, ACS Nano, 2013, 7, 11227–11233 CrossRef CAS PubMed.
- M. Kanada, M. H. Bachmann, J. W. Hardy, D. O. Frimannson, L. Bronsart, A. Wang, M. D. Sylvester, T. L. Schmidt, R. L. Kaspar and M. J. Butte, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E1433–E1442 CAS.
- W. Xue, J. E. Dahlman, T. Tammela, O. F. Khan, S. Sood, A. Dave, W. Cai, L. M. Chirino, G. R. Yang and R. Bronson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E3553–E3561 CrossRef CAS PubMed.
- L. Cecilia, E. Maria and L. Jan, J. Visualized Exp., 2012, 59, e3037 Search PubMed.
- R. T. Davies, J. Kim, S. C. Jang, E.-J. Choi, Y. S. Gho and J. Park, Lab Chip, 2012, 12, 5202–5210 RSC.
- S. M. Santana, M. A. Antonyak, R. A. Cerione and B. J. Kirby, Biomed. Microdevices, 2014, 16, 869–877 CrossRef CAS PubMed.
- K. Lee, H. Shao, R. Weissleder and H. Lee, ACS Nano, 2015, 9, 2321–2327 CrossRef CAS PubMed.
- Z. Wang, H.-j. Wu, D. Fine, J. Schmulen, Y. Hu, B. Godin, J. X. Zhang and X. Liu, Lab Chip, 2013, 13, 2879–2882 RSC.
- H. Shao, J. Chung, K. Lee, L. Balaj, C. Min, B. S. Carter, F. H. Hochberg, X. O. Breakefield, H. Lee and R. Weissleder, Nat. Commun., 2015, 6, 6999 CrossRef CAS PubMed.
- F. Wei, J. Yang and D. T. W. Wong, Biosensors and Bioelectronics, 2013, 44, 115–121 CrossRef CAS PubMed.
- Y. Yoshioka, N. Kosaka, Y. Konishi, H. Ohta, H. Okamoto, H. Sonoda, R. Nonaka, H. Yamamoto, H. Ishii and M. Mori, Nat. Commun., 2014, 5, 3591 Search PubMed.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Lab Chip, 2014, 14, 1891–1900 RSC.
- M. Jørgensen, R. Bæk, S. Pedersen, E. K. Søndergaard, S. R. Kristensen and K. Varming, J. Extracell. Vesicles, 2013, 2, 20920 Search PubMed.
- G. Kim, C. E. Yoo, M. Kim, H. J. Kang, D. Park, M. Lee and N. Huh, Bioconjugate Chem., 2012, 23, 2114–2120 CrossRef CAS PubMed.
- C. Chen, B.-R. Lin, H.-K. Wang, S.-T. Fan, M.-Y. Hsu and C.-M. Cheng, Microfluid. Nanofluid., 2014, 16, 849–856 CrossRef CAS.
- M. He, J. Crow, M. Roth, Y. Zeng and A. K. Godwin, Lab Chip, 2014, 14, 3773–3780 RSC.
- S. Ibsen, A. Sonnenberg, C. Schutt, R. Mukthavaram, Y. Yeh, I. Ortac, S. Manouchehri, S. Kesari, S. Esener and M. J. Heller, Small, 2015, 11, 5088–5096 CrossRef CAS PubMed.
- A. Sonnenberg, J. Y. Marciniak, E. A. Skowronski, S. Manouchehri, L. Rassenti, E. M. Ghia, G. F. Widhopf, T. J. Kipps and M. J. Heller, Electrophoresis, 2014, 35, 1828–1836 CrossRef CAS PubMed.
- K. Kei, K. Masashi, H. Nami, A. Takanori, K. Nobuyoshi, O. Takahiro and I. Takanori, Jpn. J. Appl. Phys, 2013, 52, 06GK10 CrossRef.
- E. J. van der Vlist, E. N. Nolte, W. Stoorvogel, G. J. Arkesteijn and M. H. Wauben, Nat. Protoc., 2012, 7, 1311–1326 CrossRef PubMed.
- V. Sokolova, A.-K. Ludwig, S. Hornung, O. Rotan, P. A. Horn, M. Epple and B. Giebel, Colloids Surf., B, 2011, 87, 146–150 CrossRef CAS PubMed.
- B. A. Ashcroft, J. de Sonneville, Y. Yuana, S. Osanto, R. Bertina, M. E. Kuil and T. H. Oosterkamp, Biomed. Microdevices, 2012, 14, 641–649 CrossRef CAS PubMed.
- C. Y. Soo, Y. Song, Y. Zheng, E. C. Campbell, A. C. Riches, F. Gunn-Moore and S. J. Powis, Immunology, 2012, 136, 192–197 CrossRef CAS PubMed.
- C. Gardiner, Y. J. Ferreira, R. A. Dragovic, C. W. G. Redman and I. L. Sargent, J. Extracell. Vesicles, 2013, 2, 19671 Search PubMed.
- C. Gercel-Taylor, S. Atay, R. H. Tullis, M. Kesimer and D. D. Taylor, Anal. Biochem., 2012, 428, 44–53 CrossRef CAS PubMed.
- A. Suetsugu, K. Honma, S. Saji, H. Moriwaki, T. Ochiya and R. M. Hoffman, Adv. Drug Delivery Rev., 2013, 65, 383–390 CrossRef CAS PubMed.
- N. Takeshita, I. Hoshino, M. Mori, Y. Akutsu, N. Hanari, Y. Yoneyama, N. Ikeda, Y. Isozaki, T. Maruyama and N. Akanuma, Br. J. Cancer, 2013, 108, 644–652 CrossRef CAS PubMed.
- D. Marimpietri, A. Petretto, L. Raffaghello, A. Pezzolo, C. Gagliani, C. Tacchetti, P. Mauri, G. Melioli and V. Pistoia, PLoS One, 2013, 8, e75054 CAS.
- K. Sugimachi, T. Matsumura, H. Hirata, R. Uchi, M. Ueda, H. Ueo, Y. Shinden, T. Iguchi, H. Eguchi and K. Shirabe, Br. J. Cancer, 2015, 112, 532–538 CrossRef CAS PubMed.
- H. Wang, L. Hou, A. Li, Y. Duan, H. Gao and X. Song, BioMed Res. Int., 2014, 2014, 864894 Search PubMed.
- C. Eichelser, I. Stückrath, V. Müller, K. Milde-Langosch, H. Wikman, K. Pantel and H. Schwarzenbach, Oncotarget, 2014, 5, 9650 CrossRef PubMed.
- B. Madhavan, S. Yue, U. Galli, S. Rana, W. Gross, M. Müller, N. A. Giese, H. Kalthoff, T. Becker and M. W. Büchler, Int. J. Cancer, 2015, 136, 2616–2627 CrossRef CAS PubMed.
- X. Huang, T. Yuan, M. Liang, M. Du, S. Xia, R. Dittmar, D. Wang, W. See, B. A. Costello and F. Quevedo, Eur. Urol., 2015, 67, 33–41 CrossRef CAS PubMed.
- S. Robert, R. Lacroix, P. Poncelet, K. Harhouri, T. Bouriche, C. Judicone, J. Wischhusen, L. Arnaud and F. Dignat-George, Arterioscler., Thromb., Vasc. Biol., 2012, 32, 1054–1058 CrossRef CAS PubMed.
- E. N. M. N.-t. Hoen, E. J. van der Vlist, M. Aalberts, H. C. H. Mertens, B. J. Bosch, W. Bartelink, E. Mastrobattista, E. V. B. van Gaal, W. Stoorvogel, G. J. A. Arkesteijn and M. H. M. Wauben, Nanomedicine, 2012, 8, 712–720 CrossRef PubMed.
- R. Lacroix, S. Robert, P. Poncelet and F. Dignat-George, Semin. Thromb. Hemostasis, 2010, 36, 807–818 CrossRef PubMed.
- D. L. M. Rupert, C. Lässer, M. Eldh, S. Block, V. P. Zhdanov, J. O. Lotvall, M. Bally and F. Höök, Anal. Chem., 2014, 86, 5929–5936 CrossRef CAS PubMed.
- R. Wyss, L. Grasso, C. Wolf, W. Grosse, D. Demurtas and H. Vogel, Anal. Chem., 2014, 86, 7229–7233 CrossRef CAS PubMed.
- D. Maiolo, L. Paolini, G. Di Noto, A. Zendrini, D. Berti, P. Bergese and D. Ricotta, Anal. Chem., 2015, 87, 4168–4176 CrossRef CAS PubMed.
- S. L. N. Maas, J. De Vrij and M. L. D. Broekman, J. Visualized Exp., 2014, 92, e51623 Search PubMed.
- R. Vaidyanathan, M. Naghibosadat, S. Rauf, D. Korbie, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, Anal. Chem., 2014, 86, 11125–11132 CrossRef CAS PubMed.
- A. Zomer, C. Maynard, F. J. Verweij, A. Kamermans, R. Schäfer, E. Beerling, R. M. Schiffelers, E. de Wit, J. Berenguer and S. I. J. Ellenbroek, Cell, 2015, 161, 1046–1057 CrossRef CAS PubMed.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz and N. Rahbari, Nature, 2015, 523, 177–182 CrossRef CAS PubMed.
- C. Lau, Y. Kim, D. Chia, N. Spielmann, G. Eibl, D. Elashoff, F. Wei, Y.-L. Lin, A. Moro and T. Grogan, J. Biol. Chem., 2013, 288, 26888–26897 CrossRef CAS PubMed.
- C. S. Hong, L. Muller, M. Boyiadzis and T. L. Whiteside, PLoS One, 2014, 9, e103310 Search PubMed.
- K. Gabriel, A. Ingram, R. Austin, A. Kapoor, D. Tang, F. Majeed, T. Qureshi and K. Al-Nedawi, PLoS One, 2013, 8, e70047 CAS.
- C. J. Beckham, J. Olsen, P.-N. Yin, C.-H. Wu, H.-J. Ting, F. K. Hagen, E. Scosyrev, E. M. Messing and Y.-F. Lee, J. Urol., 2014, 192, 583–592 CrossRef CAS PubMed.
- E. Zeringer, M. Li, T. Barta, J. Schageman, K. W. Pedersen, A. Neurauter, S. Magdaleno, R. Setterquist and A. V. Vlassov, World J. Methodol., 2013, 3, 11 CrossRef PubMed.
- K. Ueda, N. Ishikawa, A. Tatsuguchi, N. Saichi, R. Fujii and H. Nakagawa, Sci. Rep., 2014, 4, 6232 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |