Zn(II) complex with 2-quinolinecarboxaldehyde selenosemicarbazone: synthesis, structure, interaction studies with DNA/HSA, molecular docking and caspase-8 and -9 independent apoptose induction†
Nenad R. Filipovića,
Snežana Bjelogrlićb,
Aleksandar Marinkovićc,
Tatjana Ž. Verbićd,
Ilija N. Cvijetiće,
Milan Senćanskif,
Marko Rodić g,
Miroslava Vujčićh,
Dušan Sladićd,
Zlatko Strikovićd,
Tamara R. Todorović*d and
Christian D. Muller*ij
g,
Miroslava Vujčićh,
Dušan Sladićd,
Zlatko Strikovićd,
Tamara R. Todorović*d and
Christian D. Muller*ij
aFaculty of Agriculture, University of Belgrade, Nemanjina 6, Belgrade, Serbia
bNational Cancer Research Center of Serbia, Pasterova 14, Belgrade, Serbia
cFaculty of Technology and Metallurgy, University of Belgrade, Karnegijeva 4, Belgrade, Serbia
dFaculty of Chemistry, University of Belgrade, Studentski trg 12-16, Belgrade, Serbia. E-mail: tamarat@chem.bg.ac.rs
eInnovation Center of the Faculty of Chemistry, University of Belgrade, Studentski trg 12-16, Belgrade, Serbia
fCenter for Multidisciplinary Research, Institute of Nuclear Sciences “Vinča”, University of Belgrade, Belgrade, Serbia
gDepartment of Chemistry, Faculty of Sciences, University of Novi Sad, Trg Dositeja Obradovića 4, Novi Sad, Serbia
hInstitute of Chemistry, Technology and Metallurgy, University of Belgrade, Njegoševa 12, Belgrade, Serbia
iLaboratoire d’Innovation Thérapeutique, UMR 7200, Faculté de Pharmacie, Université de Strasbourg, 67401 Illkirch, France. E-mail: cdmuller@unistra.fr
jPlateforme eBioCyt, Faculté de Pharmacie & Féderation Translationnelle de Médecine, Université de Strasbourg, 67401 Illkirch, France
First published on 16th October 2015
Abstract
A new Zn(II)-based potential chemotherapeutic agent was synthesized from the ligand 2-quinolinecarboxaldehyde selenosemicarbazone (Hqasesc). Single crystal X-ray diffraction analysis showed that the Zn(II) complex consists of a cation [Zn(Hqasesc)2]2+, two perchlorate anions and one ethanol solvent molecule. The interaction of calf thymus (CT) DNA and human serum albumin (HSA) with the Zn(II) complex was explored using absorption and emission spectral methods, and also has been supported by molecular docking studies. The complex has more affinity to minor DNA groove than major, with no significant intercalation. The HSA interaction studies of the complex revealed the quenching of the intrinsic fluorescence of the HSA through a static quenching mechanism. The antitumor activity of the ligand and the complex against pancreatic adenocarcinoma cell line (AsPC-1) and acute monocytic leukemia (THP-1) cells was evaluated. Both compounds are strong concentration-dependent apoptosis inducers in THP-1 cells. While Hqasesc in AsPC-1 cells induces apoptosis only at the highest concentration, treatment with the Zn complex shows a concentration-dependent apoptotic response, where the treated cells are arrested in the G1-to-S phase accompanied with extensive activation of caspase-8 and -9. These results indicate that the ligand and Zn(II) complex display cell phenotype specific activity.
Introduction
Thiosemicarbazones are condensation products of thiosemicarbazide and carbonyl compounds, which have been extensively studied as NS bidentate ligands in coordination chemistry. They possess imine nitrogen and sulfur donor atoms from the thiosemicarbazide moiety.1,2 The coordination capacity of this class of ligands may be extended if there is a donor atom(s) from the carbonyl compound in a position(s) suitable to form a chelate ring(s). This class of compounds is well known to possess a wide range of pharmacological properties.3 Studies from the 1960s indicated that the mechanism of action of thiosemicarbazones involves the inhibition of DNA synthesis.4 When it was proven that tumor cells lose their ability to convert ribonucleotides to deoxyribonucleotides after treatment with N-heteroaromatic thiosemicarbazone obtained from 2-formylpyridine, ribonucleotide reductase (RR) was marked as the main target for NNS chelating thiosemicarbazones. Chelation of iron from the RR active site was proposed to be the reason for enzyme inactivation.4 Recent findings indicate other mechanisms of thiosemicarbazone action: reactive oxygen species (ROS) generation, DNA binding, topoisomerase II inhibition, mitochondria disruption, inhibition of protein synthesis and multidrug resistance protein inhibition.4–6Despite the fact that various thiosemicarbazones have been developed and even patented as pharmaceuticals,7 the synthesis and biological activity investigation of this type of compounds is still a topic of interest. Some recent studies resulted in new thiosemicarbazone molecules, which are more active than standard chemotherapeutics.8–13 One strategy for the improvement of biological activity of thiosemicarbazones is the synthesis of their metal complexes. Coordination of thiosemicarbazones to metals enhances their lipophilicity by orienting the lipophilic and aromatic parts outwards from the complex molecules toward the solvent.6 Metal complexes may be a vehicle for the activation of thiosemicarbazones by increasing their ability to diffuse through the semipermeable membrane of cells, and complexes often show less side effects and different bioactivities in comparison to metal-free ligands.3
Selenosemicarbazones are selenium analogues of thiosemicarbazones and their biological activity has been studied in a lesser extent, probably because selenium was considered as a toxic element for humans up to the late 1950s.14 Another reason might be the instability of this type of compounds as is apparent from the precipitation of elemental selenium during the complexation of selenosemicarbazones, modification of ligands with the evolution of hydrogen selenide, or oxidation with the formation of diselenide bridges.14 However, some of synthesized selenosemicarbazones and their complexes showed good biological activity.15–18 A few comparative studies indicated that selenosemicarbazones showed similar or even better biological activities than the corresponding thiosemicarbazones and their complexes.19–24 We previously synthesized and characterized Zn(II) complexes with N-heteroaromatic selenosemicarbazones of 2,6-diacetylpyridine, 2-formylpyridine and quinoline-2-carboxaldehyde (H2dapsesc, Hfpsesec and Hqasesc, respectively).25–27 In the corresponding complexes the ligands were coordinated via Naromatic, Nimin, and Se atoms. The Zn complexes with pyridine derivatives were the subject of antitumor investigations,27–31 while the complex with a quinoline derivative was unsuitable due to low solubility. The complex with H2dapsesc showed a strong dose-dependent cytotoxic activity, with an IC50 value in the range 13–60 μM on six human carcinoma cell lines, as determined by a colorimetric assay after 24 h incubation.30 On human breast cancer (MDA-361) and human colon cancer (LS174) cell lines, the IC50 values for this complex were of the same order of magnitude as IC50 values of cisplatin (CDDP). The Zn complex with a formylpyridine derivative showed a strong cytotoxic effect on the five treated human cancer cell lines, in the range of the activity of CDDP, as determined by a colorimetric assay after 48 and 72 h of incubation.27 It is worth mentioning that this complex had a low toxicity on normal endothelial cells (EA.hy926), lower in comparison to the activity on treated cancer cell lines, and even lower than the activity of CDDP on EA.hy926 cells.
In order to continue our investigation on the antitumor activity of selenosemicarbazone complexes with biometals we have synthesized a new Zn complex with the Hqasesc ligand and evaluated ligand and complex activities on a different cancer cell model than used in our previous studies. While organizing current research, our guiding idea was to test whether these compounds can display full-scale anticancer activity. Extensive research of cancer biology and behavior has revealed that the same cells, which mediate tumor growth, are also responsible for resistance to treatment and tumor relapse, and these have been referred as Cancer Stem Cells (CSCs).32 An important feature of CSCs is their extensive genomic heterogeneity, which results in persistent cancer sub-clones with a unique genomic and epigenomic profile, and each of these sub-clones is probably endowed with a different drug-resistant phenotype. Furthermore, it was shown that another characteristic of CSCs is the reversible transition between epithelial and mesenchymal states in carcinoma. The epithelial–mesenchymal transition (EMT) is a physiological process that occurs during embryonic development, which is defined by biochemical changes in epithelial cells that lead to the acquisition of mesenchymal properties, including loss of cell adhesion and increased migratory capacity.33 In cancer, the EMT generates cells with apparent CSC features and has been associated with more aggressive tumors.34 Therefore, evidence from experimental models and clinical studies strongly indicate that the major goal of the current drug development should be to create an efficient strategic drug with ability to induce apoptotic death in all tumor cells including CSCs, as a way to achieve a cancer cure. For that reason, in the present work we have explored the activity of the Hqasesc ligand and its Zn complex against pancreatic adenocarcinoma cell line (AsPC-1), which possesses characteristics of epithelial–mesenchymal transition (EMT)-type cells, bearing resistance to three conventional chemotherapeutic agents (gemcitabine, 5-fluorouracil, and CDDP).35,36 Antitumor activity of the compounds was also evaluated on non-CSC acute monocytic leukemia (THP-1) cells. Owing to the noted characteristics, the chosen cell lines represent excellent in vitro models for the investigation of the Hqasesc ligand and its Zn complex against the heterogeneity and plasticity of cancer cells.
Results and discussion
The Zn(II)-complex with the ligand Hqasesc was prepared by a template reaction starting from zinc(II) perchlorate hexahydrate, 2-quinolinecarboxaldehyde and selenosemicarbazide (mole ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, respectively). The complex is soluble in MeOH, EtOH, MeCN, DMF and DMSO at room temperature. Molar conductivity measurements showed that the complex is a 1
2, respectively). The complex is soluble in MeOH, EtOH, MeCN, DMF and DMSO at room temperature. Molar conductivity measurements showed that the complex is a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 electrolyte. Based on these results, as well as results of elemental analysis, spectroscopic and X-ray diffraction study (vide infra), the formula of the complex can be written as [Zn(Hqasesc)2](ClO4)2·EtOH (1).
2 electrolyte. Based on these results, as well as results of elemental analysis, spectroscopic and X-ray diffraction study (vide infra), the formula of the complex can be written as [Zn(Hqasesc)2](ClO4)2·EtOH (1).
IR and NMR spectroscopy
The experimental IR spectrum of 1 (Fig. S1a, ESI†) was interpreted in terms of a calculated one (Fig. S1b, ESI†) at the B97D/TZVP level of theory (see Experimental section for details) and with comparison to the spectrum of the metal-free ligand. In the IR spectrum of Hqasesc there are sharp medium intensity bands in the region 3340–3080 cm−1 which are assigned to the ν(NH2) and ν(NH) stretching vibrations.26 The lack of a large systematic shift of these bands in the IR spectrum of 1 indicates that there is no interaction between the terminal nitrogen atom and zinc ion. As had been previously observed in other selenosemicarbazone complexes,25–28 in the IR spectrum of 1 there is a systematic shift of the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) band to a higher frequency in comparison to that in the corresponding ligand (1607 cm−1 in Hqasesc; 1617 cm−1 in 1). Also, in the spectrum of 1 a sharp and strong band at ∼1100 cm−1 originating from the perchlorate ion can be observed.
N) band to a higher frequency in comparison to that in the corresponding ligand (1607 cm−1 in Hqasesc; 1617 cm−1 in 1). Also, in the spectrum of 1 a sharp and strong band at ∼1100 cm−1 originating from the perchlorate ion can be observed.
1D (1H and 13C) and 2D (COSY, NOESY, 1H-13C HSQC and 1H-13C HMBC) spectra of 1 were recorded in DMSO-d6 and are shown in Fig. S2–S7 (ESI†) together with the atom numbering scheme (Scheme S1, ESI†). NMR spectral data for Hqasesc are in full agreement with previously published data.26 In all the 1D spectra of 1 there is only one set of signals, indicating the presence of just one form of the complex in solution. Based on the presence of the H–N3 signal at 12.56 ppm in the 1H NMR spectrum of 1, the ligand is coordinated to Zn(II) in the neutral form. It has already been documented that NMR signal shifts for Zn-thio(seleno)semicarbazone complexes in comparison to corresponding signals for free ligands are less pronounced than for other chalcogensemicarbazone d-metal complexes.25,27,37,38 Coordination of the Zn(II) ion to Hqasesc gives rise to downfield shifts of all protons in the 1H NMR spectrum (Fig. S8a, ESI†), together with shifts of almost all signals in the 13C NMR spectrum (Fig. S8b, ESI†). The quinoline nitrogen atom coordination is corroborated by a significant downfield shift of the signal of C4–H in the proton NMR and the signal of C4 in the carbon NMR spectrum, as well as the upfield shift of signals of C8a and C2 in the carbon NMR spectrum of 1. The downfield shift of the C9 signal in the 13C NMR spectrum indicates imine nitrogen coordination, while due to selenium atom coordination the signals of N4Ha and N4Hb are significantly shifted downfield. A shift of the signal of the selenone carbon atom (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Se) in the spectrum of 1 is similar to those reported for analogous Zn-chalcogensemicarbazone complexes.25,27,37 The complex 1 was quite stable, as can be seen from the proton spectra of a freshly prepared sample and a sample after 24 h (Fig. S9, ESI†).
Se) in the spectrum of 1 is similar to those reported for analogous Zn-chalcogensemicarbazone complexes.25,27,37 The complex 1 was quite stable, as can be seen from the proton spectra of a freshly prepared sample and a sample after 24 h (Fig. S9, ESI†).
Cyclic voltammetry
The redox behavior of Hqasesc and 1 have been investigated by cyclic voltammetry (Fig. S10, ESI†) in a potential range from +1.2 to −2.0 V relative to a Ag/AgCl reference electrode. Repeated scans, as well as different scan rates, showed that dissociation of the ligand does not take place in 1. In the cyclic voltammogram of the ligand, two cathodic peaks at −1.38 and −1.93 V vs. Ag/AgCl were observed. The first peak can be attributed to imine bond reduction since a similar value (−1.33 V) was assigned to a pyridinium analogue of Hqasesc.39 It is reasonable to assume that the second peak resulted from selenoamide reduction. Namely, similar values for benzil bis(thiosemicarbazone) (−1.440 and −1.746 V vs. Ag/AgCl) were assigned to imine and thioamide reduction, respectively.40 In the reverse scan process, no anodic peaks were observed, indicating that these electrochemical processes were coupled with a chemical reaction (EC mechanism).41 In the cyclic voltammogram of 1 the corresponding cathodic peaks are shifted to positive positions (−1.35 and −1.84 V vs. Ag/AgCl) due to the metal–ligand orbital interactions, as was noticed for the Zn complex with 4-phenyl-1-[(4-methoxyphenyl)methylene]-thiosemicarbazide.42 In the range from 0 to −1.3 V additional reduction peaks are observed in the cyclic voltammogram of 1, as in the case of the Zn–benzil bis(thiosemicarbazone) complex.40 The number of reduction peaks is equal for Hqasesc and 1 which indicates that the complex is stable – the Zn(II) ion is “hidden” for reduction in the chelate structure. On the other hand, the number of oxidation peaks in the cyclic voltammograms of Hqasesc and 1 is different (Hqasesc: 0.32 and −0.02 V vs. Ag/AgCl; 1: 0.30, −0.04 and −0.52 V vs. Ag/AgCl). The additional oxidation peak can be assigned to Zn(II) ion oxidation, which is proven by addition of Zn(ClO4)2·6H2O to a solution of 1 (Fig. S10, ESI†).Determination of acidity constants of Hqasesc
The acidity constants of the ligand were determined using spectrophotometric titration at T = 298 ± 1 K. Within the studied pH range (1.65–11.91) Hqasesc acts as a sparingly soluble ampholyte (Scheme 1).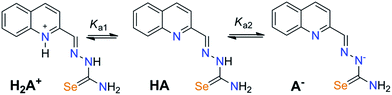 | ||
| Scheme 1 Proteolytic equilibria of Hqasesc in aqueous media. HA, as neutral form of Hqasesc, is represented in one tautomeric form, while all tautomers coexist in solution. | ||
UV-Vis spectra of Hqasesc in pH ranges 1.65–5.17 and 5.17–11.91 are shown in Fig. S11a and S12a (ESI†). The existence of two pH regions with clearly visible isosbestic points indicates that the two pKa values are well separated (ΔpKa > 3). The existence of isosbestic points proves that the only process in solution within both pH ranges is the proteolytic equilibrium shown in Scheme 1. This makes the spectrophotometric method valid for pKa value determination. Acidity constant (Ka1 and Ka2) values were calculated according to transformed forms of the classical spectrophotometric equation,43 which gives a linear dependence (for details see Experimental part). A value of Ka1 was obtained as the slope from the corresponding linear dependencies at four wavelengths (287, 327, 341, 390 nm), while Ka2 was obtained as the slope from the corresponding linear dependencies at three wavelengths (266, 330, 365 nm). Examples are shown in Fig. S11b and S12b (ESI†). The obtained pKa1 and pKa2 values are given in Table 1 and the corresponding distribution diagram in Fig. 1. It should be noted that all determined constants represent macro ionization constants. As shown in Scheme 1, Ka1 represents the dissociation of the protonated quinoline nitrogen, while Ka2 accounts for different contributions of the selenone tautomer (shown in Scheme 1) and selenenol tautomer (not shown).
| Wavelength (nm) | pKa1 | Wavelength (nm) | pKa2 |
|---|---|---|---|
| 287 | 3.52 | 266 | 7.16 |
| 327 | 3.43 | 330 | 7.18 |
| 341 | 3.40 | 365 | 7.08 |
| 390 | 3.42 | ||
| Average ± CI | 3.44 ± 0.08 | 7.14 ± 0.13 |
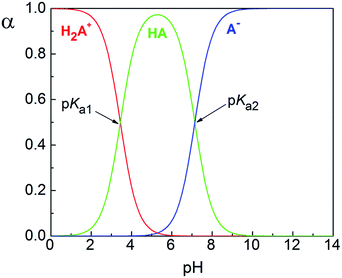 | ||
| Fig. 1 Hqasesc distribution diagram. Species notation as in Scheme 1. | ||
Description of the solid-state structures
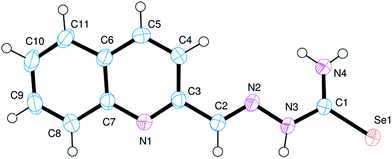
A summary of data collection and refinement parameters for Hqasesc and 1 is given in Table 2. The molecular structure of Hqasesc is presented in Fig. 2, and selected bond lengths and bond angles are presented in Table 3.| Hqasesc | 1 | |
|---|---|---|
| Molecular formula | C11H10N4Se | C24H26Cl2N8O9Se2Zn |
| Formula weight | 277.19 | 864.72 |
| Temperature (K) | 294(2) | 294(2) |
| Habitat | Irregular | Block |
| Color | Orange | Red |
| Crystal size (mm3) | 0.47 × 0.37 × 0.21 | 0.70 × 0.54 × 0.30 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P21/n | P21/c |
| a (Å) | 8.19022(13) | 11.14856(17) |
| b (Å) | 11.07458(14) | 13.88140(18) |
| c (Å) | 12.09766(18) | 20.8541(3) |
| β (°) | 92.6837(13) | 98.5190(15) |
| V (Å3) | 1096.09(3) | 3191.73(8) |
| Z | 4 | 4 |
| Dcalc (Mg m−3) | 1.68 | 1.80 |
| μ (mm−1) | 3.402 | 3.281 |
| θ range (°) | 3.0–29.0 | 3.1–32.5 |
| Reflections collected | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 948 948 |
55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 626 626 |
| Independent reflections, Rint | 2714, 0.023 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890, 0.021 890, 0.021 |
| Reflections with I > 2σ(I) | 2368 | 8743 |
| Parameters, restraints | 157, 0 | 435, 9 |
| Goodness-of-fit on F2 | 1.206 | 1.072 |
| R, wR (I > 2σ(I)) | 0.025, 0.034 | 0.037, 0.052 |
| R, wR (all data) | 0.075, 0.104 | 0.102, 0.109 |
| Δρ max and min (e Å−3) | 0.47, −0.31 | 0.84, −0.59 |
| Hqasesc | 1 | |
|---|---|---|
| Zn1–Se1 | — | 2.6233(3) |
| Zn1–Se1A | — | 2.5592(3) |
| Zn1–N1 | — | 2.2211(17) |
| Zn1–N1A | — | 2.2177(17) |
| Zn1–N2 | — | 2.1971(18) |
| Zn1–N2A | — | 2.1808(18) |
| C2–N2 | 1.278(3) | 1.275(3) |
| C2A–N2A | — | 1.271(3) |
| N2–N3 | 1.364(3) | 1.355(3) |
| N2A–N3A | — | 1.362(3) |
| C1–N3 | 1.348(3) | 1.339(3) |
| C1A–N3A | — | 1.340(3) |
| C1–Se1 | 1.841(2) | 1.842(3) |
| C1A–Se1A | — | 1.842(2) |
| N1–Zn1–N2 | — | 73.74(7) |
| N1A–Zn1–N2A | — | 74.02(7) |
| N2–Zn1–Se1 | — | 77.84(5) |
| N2A–Zn1–Se1A | — | 79.44(5) |
| N1–Zn1–Se1 | — | 149.70(5) |
| N1A–Zn1–Se1A | — | 152.98(5) |
Validation of the Hqasesc structure with MERCURY CSD44 shows no significant deviation from the common values found in the Cambridge Structural Database.45 Thus, the structural parameters of Hqasesc are quite usual for this arrangement of sp2 hybridized atoms. It is evident that electron delocalization is pronounced throughout the whole ligand molecule, and only the C2–N2 distance corresponds to a localized double bond. It is interesting to note that the ligand is comprised of two nearly planar parts – the quinoline aldehyde moiety and the selenosemicarbazide moiety – which are mutually inclined at 20.38(10)°. Moreover, the molecule adopts a conformation in which the quinoline N1 and azomethine N2, as well as the azomethine N2 and Se1 atoms are mutually in trans positions. This implies that conformational rearrangement is necessary for metal binding in the expected and observed tridentate way. Molecular arrangement in the crystal structure of the ligand is governed by a network of hydrogen bonds formed between N4 and N1 and N3 and Se1 (Fig. S13 and Table S1, ESI†) so that molecules are grouped into sheets parallel to the (−101) crystallographic plane.
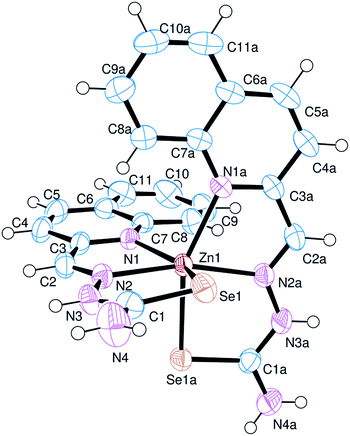
The asymmetric part of the unit cell in the crystal structure of 1 is comprised of the complex cation [Zn(Hqasesc)2]2+ (Fig. 3), two perchlorate anions and one ethanol molecule. Important structural parameters are collected in Table 3. The complex cation has approximately C2 symmetry (pseudo-symmetry axis is the bisector of N1–Zn1–N1A angle) and is comprised of two meridionally placed ligand molecules chelating the Zn(II) atom. Hqasesc molecules are coordinated in their neutral forms as tridentate NNSe donors, through the quinoline and azomethine nitrogen atoms, and selenium atom. This coordination mode of the ligand is already observed in Ni(II) and Co(III) complexes of the formula [Ni(qasesc)2]·DMSO26 and [Co(qasesc)2]BF4·2H2O,46 and as expected, it is analogous to NNS coordination of structurally related thiosemicarbazones.47–49 In that way, each ligand forms two five-membered metallocycles and all show a high degree of planarity. The exception is five-membered metallocycle Se1–Zn1–N2–N3–C1, which is significantly puckered, and may be described as twisted on the Zn1–N2 bond. The coordination octahedron is distorted, as a consequence of constraints implied by the ligand’s bite angles which are in the range 73.74(7)–79.44(5)°.
Metal–ligand bonds fall in a wide range (2.1808(18)–2.6233(3) Å), which is in line with the different covalent radii of nitrogen and selenium donor atoms. Quinoline nitrogen atoms form slightly longer bonds than azomethine nitrogen atoms, which is also observed in Ni(II) and Co(III) complexes with Hqasesc.26,46 On the other hand, significantly longer bonds are formed between Se and Zn atoms, as expected. It is worth mentioning that Zn–Se bonds are not equivalent, and that the Se1 atom forms an almost 0.1 Å longer bond [2.6233(3) Å] compared to Se1A [2.5592(3) Å]. Moreover, the Se1 atom is part of the only significantly puckered metallocycle. Ligand deformation occurs in a way that the distance between Se1 and Se1A is maximized (Se1⋯Se1A = 3.8831(3) Å). This can be visualized by torsion angles τ(C2–N2–N3–C1) = −169.7(2)°, and τ(N2–N3–C1–Se1) = −6.9(3)°. Similar behavior is observed in [Ni(qasesc)2], where the Se⋯Se distance is 3.7955(5) Å however, in that complex, both ligands are equally deformed, whereas in [Zn(Hqasesc)2]2+ only one ligand is deformed. A proposed plausible explanation is that the that bis-ligand structure of the cation places two selenium atoms closer than the sum of their van der Waals (vdW) radii (rSe = 1.9 Å), and distortion is the consequence of steric repulsion.26 Nevertheless, in [Co(qasesc)2]BF4·2H2O and Fe(III) complex [Fe(L)2][FeCl4]·1/3MeCN (HL = 3-azabicyclo[3.2.2]nonane-3-carboselenoic acid, [1-(2-pyridinyl)ethylidene]hydrazide)50 the Se⋯Se distances [3.349(1) and 3.494(3) Å, respectively] are significantly less than the sum of their vdW radii. The comparison of M–Se bond lengths and Se⋯Se distances above mentioned shows that these properties are approximately in direct proportion. Since Se⋯Se distances in Co(III) and Fe(III) complexes are shorter than the sum of the vdW radii, it appears that in these cases the strength of the coordination bonds compensate the steric repulsion, whereas in Ni(II) and Zn(II) complexes the longer metal–ligand bonds allow deformation of the coordination polyhedron to minimize steric repulsion between non-bonded adjacent Se atoms.
Intra-ligand bond lengths in the structure of 1 are quite similar to those found in the free ligand. A CSD search for C–Se single and double bond length distribution in Se-coordinated compounds having a N–C(–Se)–N fragment, reveals very broad and overlapped ranges: 1.81–1.93 Å (67 hits, mean = 1.87(2) Å) for formal double bonds, and 1.81–1.97 Å (58 hits, mean = 1.88(3) Å) for formal single bonds. In this context, Se1–C1 and Se1A–C1A bond lengths in the complex (1.842(3) and 1.842(2) Å, respectively) are slightly shorter than the average C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Se double bond in compounds with structurally related fragments.
Se double bond in compounds with structurally related fragments.
Overall the crystal structure is comprised of interleaved cation sheets and anion + solvent sheets, progressing parallel to the ac plane (Fig. S14, ESI†). In addition to the electrostatic attraction, cations and perchlorate anions are connected with numerous hydrogen bonds. The two selenosemicarbazone moieties of a cation are differently involved in these interactions as hydrogen bond donors. Namely, N(4)H2 and N(3)H groups are bonded exclusively to perchlorate oxygen atoms, whereas N(4A)H2 and N(3A)H groups are bonded to ethanol oxygen, while the N(4A)H2 group is additionally bonded to another perchlorate anion. The only significant cation–cation interaction within the sheet is the weak N4A–H4D⋯Se1iii (iii: −x, y, −1/2) hydrogen bond (Table S1, ESI†). The hydroxyl group of the ethanol molecule exhibits positional disorder, and in both sites (O9A and O9B) it acts as a N(3A)H hydrogen bond acceptor, but acts as a hydrogen bond donor to perchlorate anions in opposite directions.
Free radical-scavenging activity
Because selenium antioxidants can prevent oxidative damage, numerous animal and clinical trials have investigated the ability of these compounds to prevent the oxidative stress that is an underlying cause of cancer. Interest in selone (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Se) antioxidants can be attributed to naturally occurring selenoneine, a compound which effectively scavenges the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical.51 In our previous study the free radical-scavenging activity of selenosemicarbazones and their metal complexes by the ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)] method were determined.14 Our results showed that Hqasesc and its isomeric analogue H8qasesc (8-quinolinecarboxaldehyde selenosemicarbazone) had an excellent ABTS cation radical-scavenging effect, which was greater than that of reference antioxidant vitamin C. Coordination to Pt(II), Pd(II), Cd(II), Co(III) and Ni(II) reduced the antioxidant activity in the corresponding complexes. However, some selenosemicarbazone complexes still showed better activity than vitamin C.14
Se) antioxidants can be attributed to naturally occurring selenoneine, a compound which effectively scavenges the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical.51 In our previous study the free radical-scavenging activity of selenosemicarbazones and their metal complexes by the ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)] method were determined.14 Our results showed that Hqasesc and its isomeric analogue H8qasesc (8-quinolinecarboxaldehyde selenosemicarbazone) had an excellent ABTS cation radical-scavenging effect, which was greater than that of reference antioxidant vitamin C. Coordination to Pt(II), Pd(II), Cd(II), Co(III) and Ni(II) reduced the antioxidant activity in the corresponding complexes. However, some selenosemicarbazone complexes still showed better activity than vitamin C.14
The proton donating ability of Hqasesc and 1 was assayed using a protocol for the determination of radical scavenging activity, the DPPH method.52 IC50 values were calculated from the plotted graph of scavenging activity against the concentrations of the samples. IC50 is defined as the total antioxidant necessary to decrease the initial DPPH radical by 50%. IC50 was calculated for all compounds based on the percentage of DPPH radicals scavenged. Ascorbic acid was used as the reference compound (positive control) with concentrations from 50 to 500 μg mL−1. These results corroborate our previous finding obtained by the ABTS method that Hqasesc has excellent free radical-scavenging activity, better than vitamin C. Coordination to Zn reduces the activity of the corresponding complex 1, but still 1 is two times more active than vitamin C.
Evaluation of pro-apoptotic activity
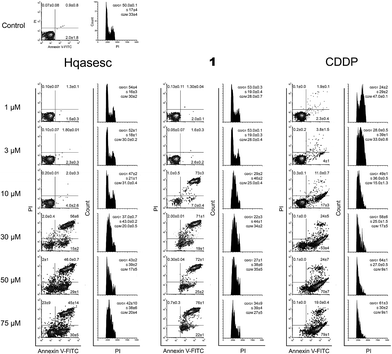
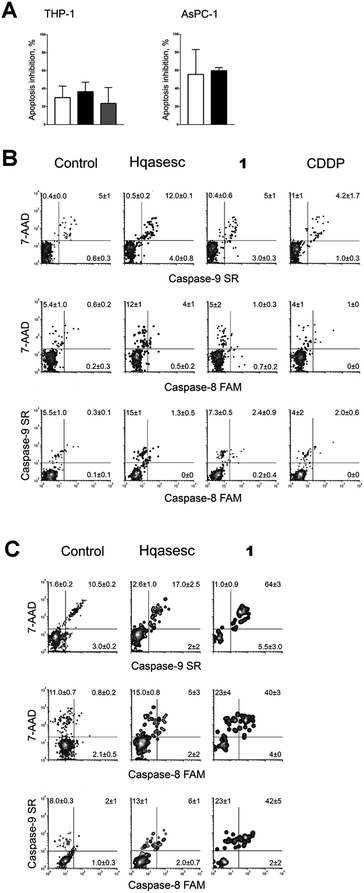
An initial experiment was organized to test the ability of Hqasesc and 1 to induce cell death in treated malignant populations. Both compounds were applied in a range of six concentrations and left for 24 h on THP-1 and AsPC-1 cells. Afterwards, cells were evaluated for the occurrence of cell death in terms of quality and quantity. There are several modalities of cell death that are classified according to morphological characteristics and measurable biochemical features.53 In the current study Annexin-V/propidium iodide (PI) staining was used, i.e. a quantitative method that provides objective assessment of cell viability alongside an accurate incidence of apoptotic and necrotic cells in the analyzed sample. This method is based on detection of phosphatidylserine (PS) externalization in the plasma membrane surface of apoptotic cells. This is the earliest event in this programmed cell death arisen as one of the “eat-me” signals that contributes to the recognition and removal of apoptotic bodies by phagocytosis.54 The PS is also a binding site for Annexin-V, while PI serves as a cell membrane non-permeant dye excluded from viable cells. Such double assay staining allows differentiation between viable cells and those in apoptosis: early (Annexin-V positive cells), late stages (Annexin-V and PI positive cells) and necrosis (PI only stained cells).As represented in Fig. 4, both compounds on THP-1 cells induced a concentration-dependent increase in percentage of apoptotic cells, while appearance of necrosis was at the level of untreated control. In cells treated with Hqasesc, apoptosis was detected starting from a concentration of 30 μM, which also was the concentration that induced the highest percent of double-stained cells. An increase of Hqasesc concentration on treated cells was followed by rise of early apoptotic events and sequential reduction of cells in late apoptosis, whereas percentages of vital cells remained almost the same in samples treated within the range 30–75 μM. Treatment with 1 induced massive apoptosis already at a concentration of 10 μM. It is interesting that at this concentration almost all apoptotic cells were in the late stages whereas the incidence of pre-apoptotic events was barely above the non-treated control. As the concentration of 1 was increased, the percentage of early apoptotic cells increased at the expense of viable cells while the incidence of double stained cells remained unchanged. Contrary to Hqasesc and 1, treatment of THP-1 cells with CDDP induced only their accumulation at the early phase of apoptosis. Even the escalation of CDDP concentration could not facilitate execution and termination of apoptosis after 24 h of incubation. It is expected from a successful anticancer agent not only to trigger apoptotic death, but also to conduct the cells through the whole process of programmed cell death. If all the chains of cascaded actions of apoptosis are accomplished within a short period of time, the treated cell will have less chance to develop resistance to the mechanism which initiated the apoptotic response. Therefore, compared to CDDP both Hqasesc and 1 displayed a more favorable mode of activity on the THP-1 cell line.
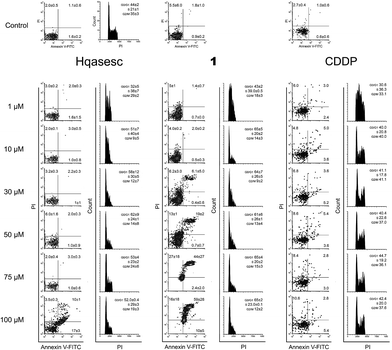
Likewise, a striking lack of early apoptotic events was also evident in AsPC-1 cells treated with 1 (Fig. 5). In these cells the first double-stained events were read-out in samples treated with 1 at 30 μM, and their percentage increased dependent on the concentration of 1. On the other hand, in AsPC-1 cells treated with Hqasesc, apoptosis was found only in the sample treated with the highest applied concentration and was not accompanied by an increased percentage of necrotic cells. These results left it unclear whether THP-1 and AsPC-1 cells, labeled with both Annexin-V and PI, after treatment with 1 were actually apoptotic or necrotic since it was shown that double-stained cells can be interpreted as both.55 In order to make a clear discrimination between apoptosis and necrosis as the endpoint event of the treatments, we decided to manipulate these two possible scenarios by using shorter incubation periods with a single appointed concentration of each compound. This concentration was chosen to be the ED50 computed according to the percentage of Annexin-V labeled cells irrespective of the PI positive cells. As seen from dose–response curves (Fig. S15, ESI†), both Hqasesc and 1 showed significantly stronger activities against THP-1 cells, with several times higher ED50 concentrations on the AsPC-1 cell line. The ED50 concentration for CDDP on THP-1 cells was comparable to the value for Hqasesc, but three times higher compared to 1. On the AsPC-1 cell line CDDP did not induce either a significant percentage of cell death, or ED50 changes.
Impact on apoptotic responses of co-incubation with the pancaspase inhibitor Z-VAD-fmk
Apoptosis is a process of biochemical events that lead to characteristic morphological changes of the cell and finally its death. These biochemical events are predetermined and include cascaded activations of different effectors, starting from a triggering incidence up to mitochondrial outer membrane permeabilization, which is marked as the “point of no return” of the lethal process.53 The central role in regulation of apoptosis belongs to caspases, a family of cysteine proteases. If caspases play an essential role in THP-1- and AsPC-1-treated cells, then the pancaspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone (Z-VAD-fmk) should inhibit the treatment-dependent response. We performed a 6 h co-incubation of Z-VAD-fmk with either Hqasesc or 1 applied at their ED50 concentrations on THP-1 and AsPC-1 cells, respectively, while the effect of Z-VAD-fmk on apoptosis induced by CDDP was investigated only on the THP-1 cell line. After incubation, the cells were stained with Annexin-V/PI, and the percentages of cell death inhibition were computed taking all Annexin-V positive events into account (Fig. 6A).According to the percent of inhibited apoptotic response for Z-VAD-fmk co-treated cells, all investigated compounds induced apoptosis by both, caspase-dependent and caspase-independent pathways. In THP-1 cells, average reduction of apoptotic death varied from 20 to 35% with no significant difference between treatments. On the contrary, the addition of Z-VAD-fmk to Hqasesc and 1 treatment on AsPC-1 cells had higher impact on cell survival as compared to THP-1 cells. These results indicated that Hqasesc and 1 induce apoptosis in THP-1 and AsPC-1 cell line by different pathways. Also, the high percent of apoptosis reduction by Z-VAD-fmk in AsPC-1 cell line treated with 1 clearly indicates that double-stained events found after 24 h of incubation were cells in advanced phases of apoptosis rather than necrosis.
Assessment of caspase activity
Results with Z-VAD-fmk imposed the need to assess which pathway is activated in regulation of apoptotic death by treatment of the cells with Hqasesc and 1. Caspases that regulate apoptosis can be divided into two major groups: caspase initiators (caspases 2, 8, 9, and 10), and caspase effectors (caspases 3, 6, and 7). Further, apoptosis can be triggered through different biochemical routes, which in general are divided into “intrinsic” and “extrinsic” pathways.56,57 Briefly, the term “extrinsic apoptotic pathway” indicates that apoptosis is triggered by extracellular death signals that are sensed and propagated by specific transmembrane receptors, a pathway regulated by caspase-8 or -10. Initiation of the “intrinsic apoptotic pathway” comes as a result of bioenergetics and metabolic catastrophe triggering caspase-9. Therefore, we evaluated caspase-8 and -9 activities after 6 h of treatment at ED50 concentrations of Hqasesc and 1 in both cell lines.Non-treated THP-1 cells show a constitutive caspase-9 activity, while activated caspase-8 was never recorded. After 6 h of treatment with Hqasesc, only a modest increase in caspase-9 activity was found whereas activity of caspase-8 stayed on the level of non treated cells (Fig. 6B). Incubation of THP-1 cells with 1 and CDDP did not induce increase any activity of these two caspases. Such a result in the case of CDDP might be quite understandable considering it induced only a small percentage of cells in the advanced phase of apoptosis, in correspondence to the percentage of apoptosis inhibited with Z-VAD-fmk co-treatment. However, as already discussed above, treatment of THP-1 cells with 1 induced a massive apoptotic response of low ED50 value with almost 40% of apoptosis inhibited by Z-VAD-fmk. All these facts indicate that there is probably another caspase-dependent pathway of apoptosis regulation involving another caspase rather than caspase-8 and -9. For instance caspase-2 should be taken into consideration. A lack of known caspase-2 substrates has been the major obstacle for its categorization within one of the two major apoptotic pathways. Caspase-2 is activated by cellular stress induced by DNA damage, cytoskeletal disruption or radical oxygen species formation, and it acts in functional compensation with caspase-9.58 Thus, further investigation will be required to identify the possible role of caspase-2 in THP-1 cell apoptosis induced by Hqasesc and 1.
Constitutive activity of caspase-9 is also found in AsPC-1 cells. Again, incubation with Hqasesc resulted in a slight increase of caspase-9 and -8 activities (Fig. 6C). On the contrary, treatment with 1 triggered massive activation of both caspases. Such a result confirms that complex 1 is a strong apoptotic inducer for AsPC-1 CSCs. Nevertheless, considerable impact of Z-VAD-fmk on apoptosis induced by Hqasesc indicates a possibility that its activity on AsPC-1 cells might also be related to the activation of caspase-2, rather than caspase-8 and -9.
Cell cycle distribution
In the current study certain improvements of the standard experimental protocols were applied. In particular, cells treated for 24 h and stained with Annexin-V/PI for evaluation of pro-apoptotic activity, after analysis on flow cytometer, were fixed overnight with ethanol and subsequently assessed for cell cycle distribution. This manipulation brought considerable advantages such as the decrease in amount of analyses, and more importantly the reduction of incidental divergences in experimental results since the same treated cells were used to provide two different but complementary read-outs.As represented in Fig. 4, Hqasesc and 1 stimulated THP-1 cells arrest in the S phase of mitotic division in a concentration-dependent manner. This phenomenon coincided with the occurrence of apoptosis. In AsPC-1 cells treatment with Hqasesc and 1 induced arrest at the S phase only at the lowest concentration of 1 μM, which, following an increase of applied concentrations, was shifted toward G1-to-S blockage (Fig. 5). Thus, the incidence of apoptosis in AsPC-1 cells could not be related to a particular alteration in cell distribution within mitotic division. Interestingly, CDDP on THP-1 cells initially induced S-to-G2 arrest at the lowest applied concentration, shifting to a blockage in the S phase at 3 μM, and finally generated G1-to-S arrest, which matches with apoptotic response (Fig. 4). These results indicate the following: Hqasesc and 1 have different modes of activity on THP-1 compared to CDDP, and Hqasesc and 1 induce apoptosis in THP-1 and AsPC-1 cells by dissimilar mechanisms. One might suppose that Hqasesc and 1 in THP-1 cells interfere with DNA replication, while in AsPC-1 cells they initiate activation of G1/S checkpoints which impedes cell transition into the S phase and the beginning of DNA replication. To determine the ability of the more active compound (1) to interact with DNA, the next set of experiments on DNA binding and DNA cleavage were performed.
DNA binding studies
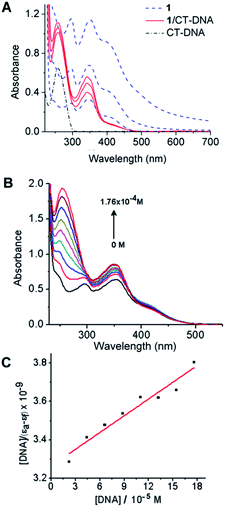
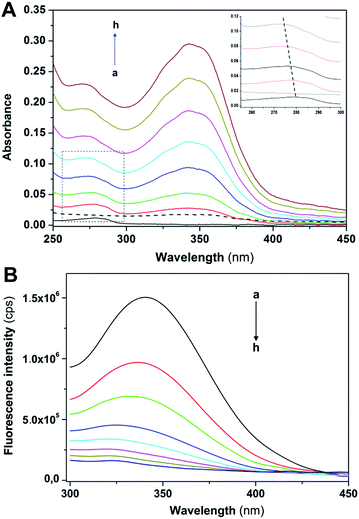
Non-covalent interactions of transition metal complexes with DNA include intercalation between the bases, binding to minor groove, major groove, sugar-phosphate backbone and three-way junction.59 Electronic absorption spectroscopy is an effective method for examining the binding mode. The hypochromic effect induced by the DNA on a spectrum of a bound ligand is one indicator of the binding mode. In general, intercalators have stronger hypochromicities than the corresponding groove binders. The absorption spectra of metal complexes bound to DNA through intercalation exhibit significant hypochromism and red shift due to the strong π → π* stacking interaction between the aromatic chromophore ligand of the metal complex and the base pairs of DNA.60 Metal complexes, which bind non-intercalatively or electrostatically with DNA, may result in either hyperchromism or hypochromism.61 Groove binding typically results in only subtle changes in the structure, and DNA remains essentially in an unperturbed B form. In contrast, intercalation in which a planar ligand moiety is inserted between adjacent base pairs, results in a substantial change in DNA structure and causes lengthening, stiffening and unwinding of the helix. Zinc-containing compounds are regarded as one of the most promising alternatives to CDDP as anticancer drugs.62 It has been reported that zinc complexes can interact with DNA by different modes.63,64 In this work, spectroscopic methods were employed to ascertain the interaction mode of 1 with calf thymus (CT) DNA.Electronic absorption spectra of 1 in aqueous buffer media, both in the absence and the presence of CT-DNA, are shown in Fig. 7. The absorption spectrum of 1 (Fig. 7A) exhibits two peaks (at 296 and 343 nm) and two shoulders (at 240–250 and 425–450 nm). The interaction of the complex with CT-DNA was monitored by the disappearance of shoulder bands and peak at 296 nm. After interaction with CT-DNA, the absorption band at 343 nm exhibited hyperchromism of about −22% with the lowest concentration of 1. With the increase of concentration of 1 (3 × 10−5 and 6 × 10−5 M), hypochromism (the value reaches 26% and 50%, respectively) followed by blue shift (10 nm at maximal performed concentration), occurs as the result of possible intercalative interactions between the complex and CT-DNA due to the strong stacking interaction between the aromatic chromophore of 1 and the base pairs of DNA.
In the spectra of Zn(II)-complex/CT-DNA, a peak at 258 nm was also observed with an increase in absorption intensity, Fig. 7A (hyperchromism was calculated as −57%, −64% and −72%). The absorption intensity at 258 nm was increased due to exposure of purine and pyrimidine bases due to complex binding. In order to obtain information on affinity of 1 for CT-DNA, a spectroscopic titration of the solution of the complex with an increasing concentration of CT-DNA was performed (Fig. 7B). The absorbance at 258 nm was monitored for each concentration of DNA. The intrinsic binding constant KB of the complex (Fig. 7C) was calculated as 8.92 × 104 M−1 (for calculation details see Experimental section). The value of binding constant was lower than reported for a classical intercalator, i.e. a range from 106 to 107 M−1.65 The value of KB for 1 was comparable to those for other metal complexes,64,66 suggesting minor groove binding of 1. The spectral changes that occur when 1 was added to CT-DNA, as well as the absence of isosbestic points in these spectra, suggest that more than a single binding mode can occur.
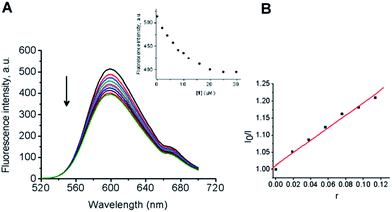
In order to further investigate the binding mode, fluorescence analyses were performed. Upon excitation at π → π* transitions either in DMSO or in the presence CT-DNA, 1 did not emit luminescence. Hence, indirect evidence for the binding mode was examined by fluorescent displacement experiments, carried out with two different dyes: ethidium bromide (EB), a typical DNA intercalator and Hoechst 33258 (H), a minor groove binder. It is known that EB interacts with CT-DNA in the intercalation mode with strong binding affinity.67 The extent of the fluorescence quenching of EB by competitive displacement from DNA is a measure of the strength of the interaction between the second molecule and DNA.68 Binding of EB to CT-DNA was followed by excitation at 500 nm with a maximum in fluorescence at 600 nm. The emission spectra of EB bound to CT-DNA in the absence and presence of 1 are given in Fig. 8. A continuous decrease was observed in fluorescence intensity at 600 nm for EB–CT-DNA with increasing concentration of 1 (up to 22 μM, see Experimental section). It was shown that the complex quenches the fluorescence probe with the maximal decrease in the fluorescence intensity of the EB–CT-DNA by 18%, followed by saturation at a high concentration of 1 (14 μM) (Fig. 8A and the inset). Fig. 8B shows that the obtained fluorescence quenching data gives a linear plot in accordance with the Stern–Volmer eqn (6) (see Experimental section). The fluorescence quenching constant (Ksv) value was calculated from the ratio of the slope to the intercept from the plot of I0/I versus r (ratio of concentrations of complex 1 and CT-DNA, r = [1]/[CT-DNA]) as Ksv = 1.802. The observed saturation at high concentrations of 1 indicated that 1 quenches the fluorescence of EB–CT-DNA by displacement of bound EB in a limited fraction of binding sites.
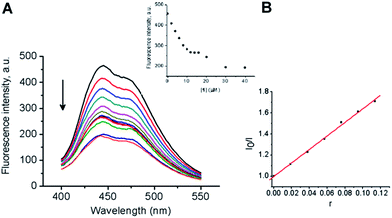
To further clarify the interaction of 1 with DNA, fluorescent displacement experiments with H dye were performed. H preferentially binds in the minor groove at A:T-rich sequences in B-DNA at the edges of four to five contiguous base pairs with nanomolar affinity.69
Also, an unspecific weaker binding at a high [H]/[DNAbp] ratio has to be considered.70 Fig. 9A shows the characteristic emission spectrum of H when it is bound to CT-DNA. The addition of 1 to CT-DNA caused appreciable reduction in the fluorescence intensity of the H–CT-DNA system in a concentration dependent way. The quenching of H–CT-DNA showed an initial saturation at 10 μM concentration of 1 (inset in Fig. 9A). It is important to note that the fluorescence intensity was reduced to nearly half of the initial value. The half-reciprocal plot of the quenching data according to the Stern–Volmer equation resulted in a linear plot with a quenching constant Ksv = 6.43. The value of quenching constant Ksv was calculated from the ratio of the slope to the intercept from the plot of I0/I versus r (r = [1]/[CT-DNA]) (Fig. 9B). With increasing concentration of 1, an additional decrease in fluorescence intensity was observed, followed by saturation in competitive displacement from the H–CT-DNA system.
The obtained results of the extent of the fluorescence quenching of EB by competitive displacement from the EB–CT-DNA system and groove binder H from the H–CT-DNA system demonstrated that 1 non-selectively binds to DNA, exhibiting both binding modes. Comparing Ksv values for the quenching fluorescence intensity by displacement it may be concluded that 1 was more efficient as a groove binder than as an intercalator. The results are in agreement with UV-Vis spectrophotometry data.
Considering that the electrostatic interaction between the positively charged complex cation [Zn(Hqasesc)2]2+ and the negatively charged phosphate backbone may occur, with respect to the geometry of the complex, it can be concluded that optimal interaction can include partial intercalation of aromatic rings in the minor groove of the double helix of CT-DNA. This mode of interaction with DNA by partial intercalation and specific minor groove binding was earlier described for bleomycins.71
Plasmid DNA interaction study
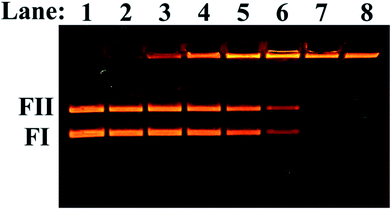
The ability of 1 to cleave double-stranded plasmid DNA in the absence of any reducing agents was investigated using an electrophoretic analysis. As is shown in Fig. 10 (lane 1), plasmid pUC19 consists mainly of supercoiled form FI and nicked form FII. DMSO did not show any effect on plasmid DNA in the performed experimental conditions (Fig. 10, lane 2). Upon addition of an increasing concentration of 1 to the plasmid, a gradual diminishing of the supercoiled form FI and nicked form FII was observed (Fig. 10, lanes 3–6) and the forms disappeared at a much higher concentration of 1 (35 and 40 μM, lanes 7 and 8, respectively). The possible multimer forms with very slow mobility were observed (remained at the start of run). The generation of the multimer was concentration dependent indicating that 1 does not possess nuclease activity. A possible cause for aggregation can be formation of multiple hydrogen bonds between the complex cation and phosphate moieties in the plasmid DNA. Such multiple hydrogen bonds were observed in the crystal structure between the complex cation and perchlorate anion, which like phosphate, has a tetrahedral geometry.Interaction between 1 and human serum albumin (HSA)
The interaction between 1 and HSA has been studied by UV-Vis and fluorescence spectroscopy. The UV-Vis spectra of HSA with increasing amounts of 1 showed a blue λmax shift (from 280 to 271 nm, Fig. 11A). The results indicate interaction between HSA and 1, leading to changes in the protein structure upon ligand binding. The changes in fluorescence emission spectra of HSA with increasing amount of 1 are shown on Fig. 11B.As the concentration of 1 increases, the results may deviate from initial linearity due to an instrumental inner filter effect.72 In order to overcome this problem, absorbance was measured at excitation (280 nm) and emission (340 nm) wavelengths, and fluorescence intensities corrected according to the Lakowicz equation73 given in the Experimental part. Corrected fluorescence intensities were used for further calculations.
Upon addition of an increasing amount of 1, a significant decrease of HSA fluorescence intensity (fluorescence quenching) is observed, as well as a blue shift of the wavelength of maximum emission (340 nm). This shift indicates that the microenvironment around Trp 214 is altered as the HSA–1 adduct is formed. In general, fluorescence quenching might be classified as static or dynamic. Dynamic quenching is highly dependent upon diffusion. Higher temperatures result in faster diffusion and hence larger values for bimolecular quenching constant. On the other hand, higher temperatures will typically result in the dissociation of weakly bound complexes, and therefore decrease the bimolecular quenching constant in a static process. Fluorescence quenching data were processed using the Stern–Volmer equation74 (for details see Experimental part) in order to obtain Ksv and Kq, the Stern–Volmer quenching constant and the quenching rate constant of the protein, respectively (Fig. S16, ESI†). As can be seen from Table 4, Ksv decreases as temperature increases, indicating that the quenching mechanism is static. Another confirmation of the static quenching mechanism is the value of Kq, which is higher than the maximum scatter collision quenching constant value (2 × 1010 M−1 s−1).75
| T (K) | 106 × Ksv (M−1) | 1014 × Kq (M−1 s−1) | r2 |
|---|---|---|---|
| 293 | 2.01 ± 0.04 | 2.82 | 0.995 |
| 298 | 1.91 ± 0.04 | 2.70 | 0.994 |
| 310 | 1.51 ± 0.03 | 2.13 | 0.993 |
The quenching process was additionally analyzed using a modified Stern–Volmer equation73 in order to obtain the effective quenching constant, Ka, for the accessible fluorophores, and the fraction of accessible fluorophore fa. The results are shown in Fig. S17 (ESI†) and in Table 5.
| T (K) | 106 × Ka (M−1) | fa | r2 | ΔH (kJ mol−1) | ΔG (kJ mol−1) | ΔS (J mol−1 K−1) |
|---|---|---|---|---|---|---|
| 293 | 1.50 ± 0.03 | 0.973 | 0.999 | −11.02 | −34.64 | 80.75 |
| 298 | 1.44 ± 0.02 | 0.972 | 0.998 | −35.13 | ||
| 310 | 1.18 ± 0.01 | 0.946 | 0.990 | −36.03 |
The Ka is inversely correlated with temperature, which is in accordance with Ksv temperature dependency.
Thermodynamic binding parameters, enthalpy (ΔH) and entropy change (ΔS), for binding of a small molecule to a protein can be calculated according to the Van’t Hoff equation (given in the Experimental section) by measuring the binding constants at several temperatures. The results are shown in Fig. S18 (ESI†) and in Table 5. According to Ross’ view,76 signs and magnitudes of thermodynamic parameters for protein reactions can account for the main forces contributing to protein stability. From the thermodynamic stand-point, ΔH > 0 and ΔS > 0 implies a hydrophobic interaction, ΔH < 0 and ΔS < 0 reflects the van der Waals force or hydrogen bond formation and ΔH < 0 and ΔS > 0 suggest an electrostatic force. In our case, ΔH < 0 and ΔS > 0, indicates that the electrostatic forces are the main contributors to HSA–1 binding.
When small molecules bind independently to a set of equivalent sites of a protein, the equilibrium between free and bound molecules for the static quenching process is given by eqn (1):77
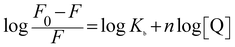 | (1) |
| T (K) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb (M−1) Kb (M−1) |
n | r2 |
|---|---|---|---|
| 293 | 5.52 ± 0.16 | 0.897 | 0.995 |
| 298 | 5.47 ± 0.18 | 0.892 | 0.994 |
| 310 | 5.73 ± 0.25 | 0.958 | 0.990 |
Fluorescence resonance energy transfer (FRET) is a non-destructive spectroscopic method that can give information about the distance and relative orientation of the donor (Trp 214, D) and acceptor fluorophore (interacting molecule, A). The resonance transfer is more efficient when spectral overlap of donor emission and acceptor absorption is large (Fig. S20, ESI†). The distance between D and A, r, could be calculated using energy transfer efficiency, E (ref. 73) (for calculation details see Experimental part). We have calculated the overlap integral of the donor emission and the acceptor absorption spectra (J = 5.87 × 10−15 M−1 cm3, Fig. S20, ESI†), the energy transfer efficiency (E = 0.230), the Förster’s distance at which energy transfer is 50% efficient (R0 = 2.385 nm), and the D–A distance (r = 3.098 nm). Since the distances between D and A are usually in the range 2–8 nm, and the obtained r value obeys the condition 0.5R0 < r < 1.5R0,73 we may conclude that the energy transfer between HSA and 1 is highly feasible process.
Molecular docking of 1 with DNA and HSA
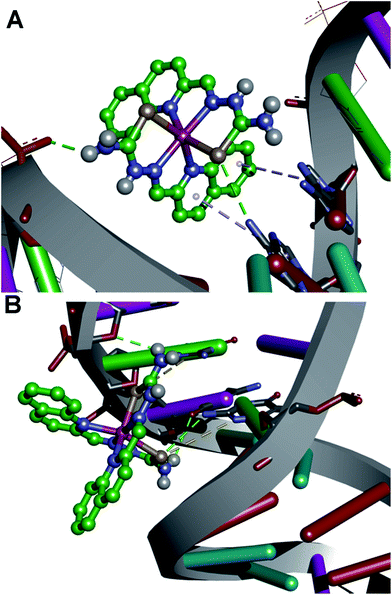
To examine the binding of the zinc complex to DNA we used the DNA duplex of sequence d(CGCGAATTCGCG)2 (for details see Experimental section). Molecular docking gave conformations binding to the minor and major groove, with slight preference for the minor groove (Fig. S21, ESI†). Conformations with lowest binding energy correspond to the minor groove, and the difference between the lowest conformation and one bound to the major groove has a value of 0.1 kcal mol−1. No intercalating structures were found. The pattern of binding is presented in Fig. 12. The established interactions between 1 and DNA nucleotides are hydrogen bonds and aromatic interactions. This is expected, as 1 consists of coordinated groups that are hydrogen bond donors, particularly NH2 groups. Aromatic systems from zinc complexes tend to form CH–π interactions with DNA bases, along with selenium (Se–π interactions), which can also form hydrogen bonds with bases. This is in accordance with the experimental results, as reported that 1 has more affinity to the minor groove than the major, and measurements showed no significant intercalation.The Zn(II) complex was docked into previously identified binding sites of HSA (PDB IDs 2BXD and 4L9Q, see Experimental part for details), but obtained values of binding energies were not in accordance with experimental results, as they were too high or showing low binding affinity. Therefore we decided to search the whole conformational space of HSA (PDB ID 1BJ5) which gave an insight into other possible binding sites. According to the reported thermodynamic properties of ligand binding, we selected one conformation that was in the best accordance with the experiment, at the same time having the lowest binding energy, with value Eb = −36.38 kJ mol−1. As presented in Fig. 13, the main electrostatic interaction is formed with Glu 354, having in mind that the complex cation is +2 charged and electrostatic effects are transmitted through organic ligands coordinated to Zn(II). Also, nitrogen atoms of Arg 209 and Lys 351 and selenium atoms are sufficiently close, so the electrostatic interaction with partially positive hydrogen atoms on the residues cannot be neglected, and this is in accordance with the reported ΔH < 0. The rest of the interactions are of hydrophobic type, CH–π and aromatic, including interaction of residues Leu 347, Phe 206, Ala 210, Val 482, Leu 481, and aliphatic chain of Lys 351 with aromatic rings of the ligand. This confirms the reported ΔS > 0 value. Also, the reported binding site is not far away from the earlier reported binding site II,78 as it is close to Trp 214 (Fig. S22, ESI†). The interactions and distances with the ligand are presented in Table S2 (ESI†). According to the docking site results, binding affinity values and interactions, it can be concluded that this can be a binding site for the reported compound, and its high affinity to HAS can be explained.
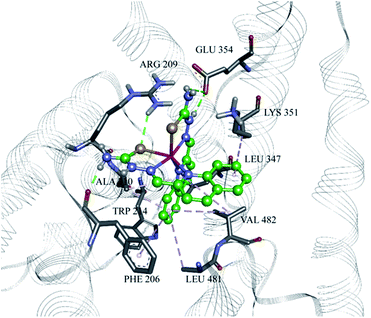 | ||
| Fig. 13 The Zn(II) complex in the binding site of HSA (PDB ID 1BJ5), with marked amino acid residues and interactions. Green: hydrogen bonds; grey: hydrophobic/aromatic interactions. | ||
Conclusions
A novel zinc(II) complex with a 2-quinolinecarboxaldehyde selenosemicarbazone ligand was synthesized and characterized. Investigation of biological activity shows that both compounds are strong apoptosis inducers in THP-1 cells. The apoptotic induction is concentration-dependent and coincides with an arrest at the S phase of mitotic division, indicating that Hqasesc and 1 interfere with DNA replication. Results of DNA binding and DNA docking experiments confirm our hypothesis, revealing that 1 not only selectively binds to DNA, but with a higher affinity for the minor groove, with no significant intercalation. Therefore, the lack of caspase-8 and -9 activation in THP-1 cells treated with 1 is consistent since apoptosis could be triggered here by caspase-2 cascade after inhibited DNA replication. On the other hand, both compounds display less activity on the AsPC-1 cell line, which one could expect since those cells are mainly CSCs, being non differentiated and so more resistant to most conventional therapeutics. While Hqasesc in AsPC-1 cells induces apoptosis only at the highest applied concentration, treatment with 1 shows a concentration-dependent apoptotic response, obviously not induced by DNA drug interaction, considering that treated cells are arrested in the G1-to-S phase, accompanied by an extensive caspase-8 and -9 activation. These results indicate that Hqasesc and 1 display cell phenotype specific activity, which is a desirable characteristic that might signify cytotoxicity on a wide spectrum of cancer types. Finally, high affinity of 1 for HSA implies a possible pharmacokinetic property. The unbound fraction of the drug in plasma may be the active form, also subjected to metabolic transformation and excretion. The HSA-bound fraction of the drug may serve as a depository, slowly releasing it as unbound active form. A high percentage of HSA-bound drug will then prolong its half-life, reducing its volume of distribution, and thus avoid possible interactions with other drugs that compete for binding to HSA.Experimental section
Reagents and instrumentation
Selenosemicarbazide (98%) was obtained from Acros Organics (BVBA, Geel, Belgium). 2-Quinolinecarboxaldehyde and zinc perchlorate hexahydrate were obtained from Aldrich (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). All solvents (reagent grade) were obtained from commercial suppliers and used without further purification.Elemental analyses (C, H, N) were performed by the standard micromethods using the ELEMENTAR Vario EL III CHNS/O analyzer. IR spectra were recorded on a Thermo Scientific Nicolet 6700 FT-IR spectrophotometer by the Attenuated Total Reflection (ATR) technique in the region 4000–350 cm−1. Abbreviations used for IR spectra: vs, very strong; s, strong; m, medium; w, weak. Molar conductivity measurements were performed at ambient temperature (298 K) on a Crison Multimeter MM41. NMR spectral assignments and structural parameters were obtained by combined use of 1H homonuclear spectroscopy (2D COSY and NOESY) and multinuclear proton-detected spectroscopy (2D HSQC, 2D HMBC). The NMR spectra were performed on a Bruker Avance 500 equipped with broad-band direct probe. Chemical shifts are given on δ scale relative to tetramethylsilane (TMS) as internal standard for 1H and 13C. Abbreviations used for NMR spectra: s, singlet; dd, doublet of doublets; ddd, double double doublet. Cyclic voltammetric measurements were performed using an electrochemical system CH Instruments (USA). UV-Vis spectra for acidity constant determination were recorded on a Thermo Scientific Evolution 60S spectrophotometer (Thermo Fisher Scientific Inc, Waltham, Massachusetts, USA) in 250–500 nm range. pH values were measured using a CRISON pH-Burette 24 2S equipped with a CRISON 50 29 micro-combined pH electrode (CRISON INSTRUMENTS, S.A. Spain). For DPPH scavenging activity, absorbance at 517 nm was measured using a Thermo Scientific Appliskan. Pro-apoptotic activity was determined on a Guava EasyCyte™ micro-capillary flow cytometer (EMD Millipore, Darmstadt, Germany) using the InCyte® software package (EMD Millipore, Cat. no. 0500-4120). UV-Vis spectra for DNA binding studies were recorded on a UV-1800 Shimadzu UV-Visible spectrometer operating from 200 to 800 nm in 1.0 cm quartz cells, while for fluorescence measurements a Thermo Scientific Lumina Fluorescence spectrometer (Finland) equipped with a 150 W xenon lamp was used. A Submarine Mini-gel Electrophoresis Unit (Hoeffer HE 33) with an EPS 300 power supply was used for DNA cleavage studies. The stained gel was illuminated under a UV transilluminator Vilber-Lourmat (France) at 312 nm and photographed with a Nikon Coolpix P340 Digital Camera through filter Deep Yellow 15 (Tiffen, USA). UV-Vis spectra for HSA binding studies were recorded on a Thermo Scientific Evolution 60S UV-Vis spectrophotometer, using a quartz cell with 1.0 cm path length, while fluorescence measurements were performed on a spectrofluorometer Fluoromax-4 Jobin Yvon (Horiba Scientific, Japan), equipped with a Peltier element for temperature control and a magnetic stirrer for cuvette, using a quartz cell with 1 cm path length.
Synthesis of Hqasesc
The ligand was synthesized according to the reported procedure.14 Single crystals suitable for X-ray analysis were obtained by recrystallization of the crude product from EtOH. The chemical structure of Hqasesc was confirmed by elemental analysis, IR and NMR spectroscopy. These data are in good agreement with the data previously reported.14Synthesis of [Zn(Hqasesc)2](ClO4)2·EtOH (1)
Into the solution of 2-quinolinecarboxaldehyde (0.15 g, 0.96 mmol) and selenosemicarbazide (0.13 g, 0.96 mmol) in EtOH (20 mL), Zn(ClO4)2·6H2O (0.18 g, 0.48 mmol) was added. The reaction mixture was refluxed for 30 min and cooled to ambient temperature. Dark red crystals of 1 were obtained from the ethanolic solution, filtered off and washed with cold ethanol and ether. Yield: 0.28 g (66%). Found: C, 33.5; H, 3.1; N, 12.9. Calc. for C24H26Cl2N8O9Se2Zn: C, 33.3; H, 3.0; N, 13.0%. IR (ATR), νmax/cm−1: 3434m (NH2), 3297vs (NH), 3183vs (NH), 3077s (NH2), 1617s (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1578s (NH), 1511m (CH), 1100vs (ClO4−). ΛM (1 × 10−3 M, EtOH): 72.0 Ω−1 cm2 mol−1. 1H NMR (500 MHz, DMSO, TMS) [ppm] δ: 12.56 (s, 1H, N3H), 9.28 (s, 1H, N4Hb), 9.14 (s, 1H, N4Ha), 8.86 (d, 1H, C4H, 3J4,3 = 8.8 Hz), 8.72 (d, 1H, C3H, 3J3,4 = 8.8 Hz), 8.38 (s, 1H, C9H), 8.19 (dd, 1H, C5H, 3J5,6 = 7.7 Hz, 4J5,7 = 1.1 Hz), 8.13 (dd, 1H, C8H, 3J8,7 = 8.5 Hz, 4J8,6 = 1.3 Hz), 8.01 (ddd, 1H, C7H, 3J7,8 = 8.5 Hz, 3J7,6 = 7.0 Hz, 4J7,5 = 1.1 Hz), 7.83 (ddd, 1H, C6H, 3J6,5 = 7.7 Hz, 3J6,7 = 7.0 Hz, 4J6,8 = 1.3 Hz). 13C NMR (126 MHz, DMSO, TMS) [ppm] δ: 175.86 (C10), 151.04 (C2), 142.42 (C4), 140.71 (C8a), 136.57 (C9), 133.28 (C7), 128.98 (C6), 128.77 (C5), 128.12 (C4a), 123.27 (C8), 119.23 (C3). Numbering of atoms used in NMR is given in Scheme S1 (ESI†).
N), 1578s (NH), 1511m (CH), 1100vs (ClO4−). ΛM (1 × 10−3 M, EtOH): 72.0 Ω−1 cm2 mol−1. 1H NMR (500 MHz, DMSO, TMS) [ppm] δ: 12.56 (s, 1H, N3H), 9.28 (s, 1H, N4Hb), 9.14 (s, 1H, N4Ha), 8.86 (d, 1H, C4H, 3J4,3 = 8.8 Hz), 8.72 (d, 1H, C3H, 3J3,4 = 8.8 Hz), 8.38 (s, 1H, C9H), 8.19 (dd, 1H, C5H, 3J5,6 = 7.7 Hz, 4J5,7 = 1.1 Hz), 8.13 (dd, 1H, C8H, 3J8,7 = 8.5 Hz, 4J8,6 = 1.3 Hz), 8.01 (ddd, 1H, C7H, 3J7,8 = 8.5 Hz, 3J7,6 = 7.0 Hz, 4J7,5 = 1.1 Hz), 7.83 (ddd, 1H, C6H, 3J6,5 = 7.7 Hz, 3J6,7 = 7.0 Hz, 4J6,8 = 1.3 Hz). 13C NMR (126 MHz, DMSO, TMS) [ppm] δ: 175.86 (C10), 151.04 (C2), 142.42 (C4), 140.71 (C8a), 136.57 (C9), 133.28 (C7), 128.98 (C6), 128.77 (C5), 128.12 (C4a), 123.27 (C8), 119.23 (C3). Numbering of atoms used in NMR is given in Scheme S1 (ESI†).
Direct syntheses of 1 starting from zinc perchlorate and Hqasesc in EtOH afforded an identical product as the template synthesis, but in a slightly lower yield.
X-ray crystallography
Single crystal X-ray diffraction was performed on an Oxford Diffraction Gemini S kappa geometry diffractometer, equipped with Mo Kα radiation from a sealed tube source, and a Sapphire CCD detector. Data collection strategy calculation, data reduction, cell refinement and absorption correction were performed with the CRYSALISPRO.79 The structure was solved using SHELXT,80 and refined with anisotropic displacement parameters for all non-hydrogen atoms using SHELXL-2014.81 Program SHELXLE82 was used as graphical user interface for structure solution and refinement procedures. Hydrogen atoms bonded to carbon atoms were introduced in idealized positions and refined using a riding model. Hydrogen atoms bonded to nitrogen atoms were located by difference Fourier synthesis, and refined using distance restraints with Uiso approximated by 1.2Ueq of the parent atoms in Hqasesc, whilst their free isotropic refinement was performed in 1. Hydroxyl group of the EtOH molecule in 1 showed positional disorder, which was modeled by refining occupational parameters, and applying distance restraints. Structure was validated using PLATON,83 and Cambridge Structural Database (CSD) (v. 5.36, updates Nov. 2014)45 using MERCURY CSD.44Crystallographic data for Hqasesc and 1 have been deposited with the Cambridge Crystallographic Data Centre as Supplementary publication no. CCDC 1413656 and 1413657,† respectively.
Cyclic voltammetry
The cell (10 mL) consisted of a three-electrode system, glassy carbon electrode (inner diameter of 3 mm; CHI 104) an Ag/AgCl (saturated KCl) reference electrode and Pt counter electrode. The potential was swept over the range from −2.0 to +1.2 V (vs. Ag/AgCl) at scan rate of 100 mV s−1. Measurements were performed at room temperature with deaeration of solutions by passing a stream of nitrogen through the solution for 5 min prior to the measurement and then maintaining a blanket atmosphere of nitrogen over the solution during the measurement. The potentials were measured in 0.10 M [n-Bu4N]PF6/DMSO, and are quoted relative to Ag/AgCl reference electrode.Determination of Hqasesc acidity constants
Acidity constants of Hqasesc were determined by spectrophotometric titration at T = 298 ± 1 K. The working solution (c = 3.1 × 10−5 M) was prepared by dissolving an accurately weighed dried substance in 5.00 mL of DMSO; 10 mM phosphate buffer (pH = 1.77, I = 0.1 M NaCl) was added up to 100.00 mL. The pH value was adjusted to 1.65 with conc. HCl. Small volume increments of 1 M KOH were added stepwise, until pH 11.91 was reached (total volume change at the end of the titration was lower than 2%). The pH value was continuously measured during the titration and UV-Vis spectra recorded after pH value equilibration. The UV-Vis spectra were recorded against the phosphate buffer as a blank. The pH electrode was calibrated by standard CRISON buffer solutions (pH 4.01, 7.00, and 9.21). Acidity constants for Hqasesc were calculated according to transformed forms of classical spectrophotometric equations43 which give linear dependence:
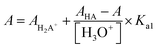 | (2) |
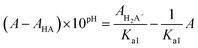 | (2a) |
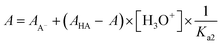 | (3) |
| (A − AHA) × 10−pH = Ka2 × AA− − Ka2 × A | (3a) |
Free radical scavenging antioxidant assay (DPPH method)
The proton donating ability was assayed using a protocol for the determination of radical scavenging activity.52 Compounds were dissolved in pure DMSO and were diluted into ten different concentrations. Commercially available free radical DPPH was dissolved in methanol at a concentration of 6.58 × 10−5 M. Into a 96-well microplate, 140 μL of DPPH solution was loaded and 10 μL DMSO solution of the tested compounds was added, or pure DMSO (10 μL) as the control. The microplate was incubated for 30 min at 298 K in the dark and the absorbance was measured at 517 nm. All the measurements were carried out in triplicate. The scavenging activity of the compounds was calculated using the eqn (4):
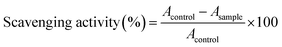 | (4) |
IC50 values were calculated from the plotted graph of scavenging activity against the concentrations of the samples. IC50 is defined as the total antioxidant concentration necessary to decrease the amount of the initial DPPH radical by 50%. IC50 was calculated for all compounds based on the percentage of DPPH radicals scavenged. Ascorbic acid was used as the reference compound (positive control) with concentrations 50 to 500 μg mL−1.
Cell cultures
Acute monocytic leukemia cell line (THP-1, ATCC® TIB-202) and pancreatic adenocarcinoma cell line (AsPC-1, ATCC® CRL-1682) were maintained in RPMI-1640 (Life Technologies, 11875-093) and DMEM high glucose (Sigma-Aldrich, D5796) cell culture media, respectively. Both media were supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, Cat. no. 16000-036) and 1% (v/v) penicillin–streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per mL and 10
000 units per mL and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg mL−1, Gibco, Cat. no. 15140-130). Cells were kept at 37 °C in an atmosphere containing 5% (v/v) CO2 during their exponential growth and during the course of experimental treatments.
000 μg mL−1, Gibco, Cat. no. 15140-130). Cells were kept at 37 °C in an atmosphere containing 5% (v/v) CO2 during their exponential growth and during the course of experimental treatments.
The ligand Hqasesc and 1 were initially dissolved in DMSO to the stock concentration of 20 mM, whereas CDDP was dissolved in saline to the stock concentration of 5 mM. Further dilutions to the experimental concentrations applied on the cells have been done with RPMI-1640 or DMEM media immediately before each experiment, thus the final concentration of DMSO with cells treated with the highest applied concentration was 0.5% (v/v).
Determination of pro-apoptotic activity
THP-1 and AsPC-1 cells were seeded in 96 flat bottom well plates (Corning® Costar®, Cat. no. CLS3596) in a volume of 0.1 mL, at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per well. Since the THP-1 cells grow in suspension, experimental treatments were started within 2 h after cell seeding, while plates with seeded AsPC-1 cells were left overnight to settle. Investigated compounds were added in a range of six concentrations. As controls, non-treated cells, cells treated with 0.5% DMSO, and cells treated with Celastrol (Enzo Life Sciences, Cat. no. ALX-350-332-M025) at 50 μM concentration were used.
000 per well. Since the THP-1 cells grow in suspension, experimental treatments were started within 2 h after cell seeding, while plates with seeded AsPC-1 cells were left overnight to settle. Investigated compounds were added in a range of six concentrations. As controls, non-treated cells, cells treated with 0.5% DMSO, and cells treated with Celastrol (Enzo Life Sciences, Cat. no. ALX-350-332-M025) at 50 μM concentration were used.
After 24 h of incubation, THP-1 cells were centrifuged at an RCF of 450 × g for 10 min, supernatants were discarded, and 0.1 mL of PBS was added to each well. Plates were placed on a plate shaker for 3 min, afterwards Annexin-V-FITC (ImmunoTools, Cat. no. 31490013) and propidium iodide (PI, MiltenylBiotec, Cat. no. 130-093-233) were added in a volume of 3 μL per well, respectively, and incubated for 15 min in the dark before cytometry analysis. Post incubation manipulation with AsPC-1 cells included trypsinization (200 μL of trypsin per well, Sigma-Aldrich, Cat. no. T3924). Subsequent steps with addition of PBS, Annexin-V and PI were the same as for THP-1 cells. Plates were analyzed on a flow cytometer. Cells were classified according to Annexin-V and PI labeling into viable (non-stained cells), pre-apoptotic cells (stained with Annexin-V only), cells in late phases of apoptosis (double stained cells), and necrotic cells (stained with PI only). After this read-out was ended, the remaining cells were fixed in ethanol overnight at 4 °C, and afterwards stained with FxCycle™ PI/RNAse Staining solution (Molecular Probes, Cat. no. F10797), and analyzed as described previously.84
Calculation of ED50 concentration
Separately for THP-1 and AsPC-1 cells, the percent of Annexin-V labeled cells for each investigated concentration of compounds were summarized and the maximum apoptotic response was normalized to 100%. The percent of apoptosis read-out for other concentrations were calculated as a proportion of the highest response. Such scaled apoptotic outcomes were plotted against concentrations and ED50 concentration was calculated using the asymmetric sigmoidal curve five-parameter logistic equation (GraphPad Prism 6 software). The determined ED50 concentrations of both compounds were applied in further steps of this investigation.Inhibition of caspase activity
THP-1 and AsPC-1 cells were treated with investigated compounds at the ED50 concentration for 6 h with or without pan-caspase inhibitor Z-VAD-fmk (Promega, Cat. no. G7232). Previously, the proper concentration of Z-VAD-fmk that is non-toxic to cells during 6 h of incubation was determined (10 μM and 20 μM for THP-1 and AsPC-1 cell lines, respectively). As controls, non-treated cells, cells treated with Z-VAD-fmk only, and cells treated with ED50 concentrations only were used. After the incubation period had ended, plates with THP-1 and AsPC-1 treated cells were stained with Annexin-V/PI as described above, and analyzed on a Guava EasyCyte™ cytometer. The percent of apoptosis inhibited by Z-VAD-fmk co-treatment was determined by the formula: apoptosis inhibition, % = [1 − (% of apoptosis in A/% of apoptosis in B)] × 100, where A is the sample treated with both ED50 and Z-VAD-fmk, while B is the corresponding sample treated with ED50 only.Evaluation of caspase-8 and -9 activities
THP-1 and AsPC1 cells were treated with the investigated compounds at the ED50 concentration for 6 h. Afterwards the activity of caspase-8 and -9 were assayed by means of a Guava Caspase 9 SR and a Caspase 8 FAM kit (EMD Millipore, Cat. no. 4500-0640), following the instructions of the manufacturer. In acquired data cells were discriminated according to expression of green fluorescence (caspase-8), yellow fluorescence (caspase-9), or red fluorescence (7-AAD) as the following: live cells (not stained with either caspase nor 7-AAD), mid-stage apoptotic cells (cells stained with either caspase 8 or 9, but negative to 7-AAD), late stage apoptotic cells (cells stained with either of caspase 8 or 9 and with 7-AAD), necrotic cells (cells not stained with either caspase 8 nor 9, but positive for 7-AAD).DNA binding experiments
Calf thymus DNA (lyophilized, highly polymerized, obtained from Serva, Heidelberg) (CT-DNA) was dissolved in Tris buffer (10 mM Tris–HCL pH 7.9) overnight at 4 °C. This stock solution (2.86 mg mL−1) was stored at 4 °C and was stable for several days. A solution of CT-DNA in water gave a ratio of UV absorbance at 260 and 280 nm, A260/A280 of 1.89–2.01, indicating that the DNA was sufficiently free of protein. The concentration of DNA (2.86 mg mL−1) was determined from the UV absorbance at 260 nm using the extinction coefficient ε260 = 6600 M−1 cm−1.85 The complex was dissolved in DMSO in concentrations of 10 mM. This solution was used as stock solution.| [DNA] × (εa − εf)−1 = [DNA] × (εb − εf)−1 + Kb−1 × (εb − εf)−1 | (5) |
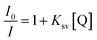 | (6) |
DNA cleavage experiments
For DNA cleavage experiments the plasmid pUC19 (2686 bp, purchased from Sigma-Aldrich, USA) was prepared by its transformation in chemically competent cells Escherichia coli strain XL1 blue. Amplification of the clone was done according to the protocol for growing E. coli culture overnight in LB medium at 37 °C (ref. 87) and purification was performed using a Qiagen Plasmid plus Maxi kit. Finally, DNA was eluted in 10 mM Tris–HCl buffer and stored at −20 °C. The concentration of plasmid DNA (512 ng μL−1) was determined by measuring the absorbance of the DNA-containing solution at 260 nm. One optical unit corresponds to 50 μg mL−1 of double stranded DNA.The cleavage reaction of supercoiled pUC19 DNA by different concentrations of 1 was investigated by incubation of 512 ng of plasmid in a 20 μL reaction mixture in 40 mM bicarbonate buffer (pH 8.4) at 37 °C, for 90 min. The reaction mixtures were vortexed from time to time. The reaction was terminated by short centrifugation at an RCF of 6708 × g and addition of 5 μL of loading buffer [0.25% bromophenol blue, 0.25% xylene cyanol FF and 30% glycerol in TAE buffer, pH 8.24 (40 mM Tris–acetate, 1 mM EDTA)]. The samples were subjected to electrophoresis on 1% agarose gel (Amersham Pharmacia-Biotech, Inc) prepared in TAE buffer pH 8.24. The electrophoresis was performed at a constant voltage (80 V) until bromophenol blue had passed through 75% of the gel. After electrophoresis, the gel was stained for 30 min by soaking it in an aqueous ethidium bromide solution (0.5 μg mL−1).
HSA binding experiments
Fatty acid free HSA (<0.007% fatty acids, Mw = 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478 Da) was purchased from Sigma, as well as potassium dihydrogen phosphate, disodium hydrogen phosphate, sodium chloride and potassium chloride used for 1× PBS preparation. A stock solution of HSA (c = 2.78 × 10−3 M) was prepared by dissolving an accurately weighed mass of commercially available lyophilized HSA in freshly prepared 1× PBS (pH = 7.35), and kept in a freezer in 100 μL portions. A stock solution of 1 (c = 6.36 × 10−4 M) was prepared by dissolving an appropriate amount of substance in DMSO. For HSA–1 interaction studies, the HSA solution was freshly prepared from the stock by dilution with PBS buffer (HSA concentration was kept constant, c = 5 × 10−7 M), and titrated with small volume increments of 1 stock solution (to avoid large sample dilution). The complex was added in 0.7, 1.4, 2.8, 4.2, 6.3, 8.4 and 10.5 molar equivalents. After each aliquot addition, the system was stirred and left to equilibrate for 15 min before UV-Vis absorption and fluorescence emission spectra recording. After the last equivalent, the total DMSO did not exceed 2% in volume. It was previously shown that addition of 15% of DMSO did not induce structural changes in bovine serum albumin, a protein structurally similar to HSA.88 Therefore, it is unlikely that the conformation of HSA was changed with the level of DMSO used in this study. Millipore water was used for all aqueous solutions. All UV-Vis spectra were recorded against the corresponding blank (PBS) in the 250–450 nm wavelength range at room temperature.
478 Da) was purchased from Sigma, as well as potassium dihydrogen phosphate, disodium hydrogen phosphate, sodium chloride and potassium chloride used for 1× PBS preparation. A stock solution of HSA (c = 2.78 × 10−3 M) was prepared by dissolving an accurately weighed mass of commercially available lyophilized HSA in freshly prepared 1× PBS (pH = 7.35), and kept in a freezer in 100 μL portions. A stock solution of 1 (c = 6.36 × 10−4 M) was prepared by dissolving an appropriate amount of substance in DMSO. For HSA–1 interaction studies, the HSA solution was freshly prepared from the stock by dilution with PBS buffer (HSA concentration was kept constant, c = 5 × 10−7 M), and titrated with small volume increments of 1 stock solution (to avoid large sample dilution). The complex was added in 0.7, 1.4, 2.8, 4.2, 6.3, 8.4 and 10.5 molar equivalents. After each aliquot addition, the system was stirred and left to equilibrate for 15 min before UV-Vis absorption and fluorescence emission spectra recording. After the last equivalent, the total DMSO did not exceed 2% in volume. It was previously shown that addition of 15% of DMSO did not induce structural changes in bovine serum albumin, a protein structurally similar to HSA.88 Therefore, it is unlikely that the conformation of HSA was changed with the level of DMSO used in this study. Millipore water was used for all aqueous solutions. All UV-Vis spectra were recorded against the corresponding blank (PBS) in the 250–450 nm wavelength range at room temperature.
Before recording the fluorescence spectrum, diluted HSA solution was ultrafiltrated using filters with 0.23 μm pore size. The excitation wavelength was 280 nm, with 5 nm slits; emission spectra were recorded in the 300–450 nm wavelength range, with 5 nm slits, and 0.1 s integration time. The background PBS signal was subtracted from each spectrum. In order to remove the instrumental inner filter effect, the absorbances were measured at the excitation (280 nm) and emission (340 nm) wavelengths, and fluorescence intensities corrected according to the Lakowicz eqn (7):73
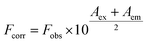 | (7) |
Fluorescence quenching data were processed using the Stern–Volmer eqn (8):73
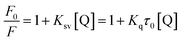 | (8) |
The quenching process was additionally analyzed using a modified Stern–Volmer eqn (9):73
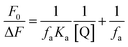 | (9) |
Thermodynamic parameters of binding, the enthalpy (ΔH) and entropy change (ΔS), during the binding of 1 to HSA were determined by measuring the binding constants at several temperatures, and following the Van’t Hoff eqn (10):
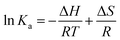 | (10) |
The distance between donor and acceptor, r, was calculated using eqn (11) for energy transfer efficiency, E:73
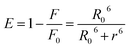 | (11) |
| R06 = 8.79 × 10−25κ2n−4ϕJ | (12) |
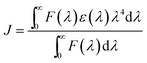 | (13) |
Computational details
Acknowledgements
The authors acknowledge networking support by the COST Action CM1106 StemChem – “Chemical Approaches to Targeting Drug Resistance in Cancer Stem Cells”. The work was founded by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grants 172055 and 172013). PARADOX computer cluster is supported in part by the Ministry of Education, Science and Technological Development of the Republic of Serbia (No. 171017) and by the European Commission under the FP7 projects (HP-SEE, PRACE-1IP, PRACE-2IP, EGI-InSPIRE).References
- T. S. Lobana, R. Sharma, G. Bawa and S. Khanna, Coord. Chem. Rev., 2009, 253, 977–1055 CrossRef CAS.
- J. S. Casas, M. S. Garcia-Tasende and J. Sordo, Coord. Chem. Rev., 2000, 209, 197–261 CrossRef CAS.
- H. Beraldo and D. Gambino, Mini-Rev. Med. Chem., 2004, 4, 159–165 Search PubMed.
- D. S. Kalinowski, P. Quach and D. R. Richardson, Future Med. Chem., 2009, 1, 1143–1151 CrossRef CAS PubMed.
- N. S. H. N. Moorthy, N. M. F. S. A. Cerqueira, M. J. Ramos and P. A. Fernandes, Recent Pat. Anti-Cancer Drug Discovery, 2013, 8, 168–182 CrossRef CAS PubMed.
- G. Pelosi, Open Crystallogr. J., 2010, 3, 16–28 CrossRef CAS.
- N. S. H. N. Moorthy, N. M. F. S. A. Cerqueira, M. J. Ramos and P. A. Fernandes, Mini-Rev. Med. Chem., 2013, 13, 1862–1872 CrossRef CAS PubMed.
- B. Shakya, P. N. Yadav, J. Ueda and S. Awale, Bioorg. Med. Chem. Lett., 2014, 24, 458–461 CrossRef CAS PubMed.
- T. A. Yousef, G. M. Abu El-Reash, O. A. El-Gammal and S. F. Ahmed, Polyhedron, 2014, 81, 749–763 CrossRef CAS.
- A. Y. Lukmantara, D. S. Kalinowski, N. Kumar and D. R. Richardson, J. Inorg. Biochem., 2014, 141, 43–54 CrossRef CAS PubMed.
- C. Gan, J. Cui, S. Su, Q. Lin, L. Jia, L. Fan and Y. Huang, Steroids, 2014, 87, 99–107 CrossRef CAS PubMed.
- A. Mrozek-Wilczkiewicz, M. Serda, R. Musiol, G. Malecki, A. Szurko, A. Muchowicz, J. Golab, A. Ratuszna and J. Polanski, ACS Med. Chem. Lett., 2014, 5, 336–339 CrossRef CAS PubMed.
- M. A. Bhat, A. Al-Dhfyan, A. A. Khan, N. Al-Harbi, P. S. Manogaran, A. M. Alanazi, H. K. Fun and M. A. Al-Omar, Bioorg. Med. Chem. Lett., 2015, 25, 83–87 CrossRef CAS PubMed.
- N. Filipović, N. Polović, B. Rašković, S. Misirlić-Denčić, M. Dulović, M. Savić, M. Nikšić, D. Mitić, K. Anđelković and T. Todorović, Monatsh. Chem., 2014, 145, 1089–1099 CrossRef.
- A. Molter, J. Rust, C. W. Lehmann, G. Deepa, P. Chiba and F. Mohr, Dalton Trans., 2011, 40, 9810–9820 RSC.
- A. Molter, G. N. Kaluđerović, H. Kommera, R. Paschke, T. Langer, R. Pöttgen and F. Mohr, J. Organomet. Chem., 2012, 70, 80–86 CrossRef.
- V. Zaharia, A. Ignat, B. Ngameni, V. Kuete, M. L. Moungang, C. N. Fokunang, M. Vasilescu, N. Palibroda, C. Cristea, L. Silaghi-Dumitrescu and B. T. Ngadjui, Med. Chem. Res., 2013, 22, 5670–5679 CrossRef CAS.
- H. Shen, H. Zhu, M. Song, Y. Tian, Y. Huang, H. Zheng, R. Cao, J. Lin, Z. Bi and W. Zhong, BMC Cancer, 2014, 14, 629–639 CrossRef PubMed.
- M. D. Revenko, V. I. Prisacari, A. V. Dizdari, E. F. Stratulat, I. D. Corja and L. M. Proca, Pharm. Chem. J., 2011, 45, 351–354 CrossRef CAS.
- K. C. Agrawal, B. A. Booth, R. L. Michaud, E. C. Moore and A. C. Sartorelli, Biochem. Pharmacol., 1974, 23, 2421–2429 CrossRef CAS PubMed.
- C. R. Kowol, R. Eichinger, M. A. Jakupec, M. Galanski, V. B. Arion and B. K. Keppler, J. Inorg. Biochem., 2007, 100, 1946–1957 CrossRef PubMed.
- C. Pizzo, P. Faral-Tello, G. Salinas, M. Flo, C. Robello, P. Wipfe and S. G. Mahler, Med. Chem. Commun., 2012, 3, 362–368 RSC.
- S. R. Turk, C. Shipman Jr and J. C. Drach, J. Gen. Virol., 1986, 67, 1625–1632 CrossRef CAS PubMed.
- V. Calcatierra, O. Lopez, J. G. Fernandez-Bolanos, G. B. Plata and J. M. Padron, Eur. J. Med. Chem., 2015, 94, 63–72 CrossRef CAS PubMed.
- T. R. Todorović, A. Bacchi, G. Pelizzi, N. O. Juranić, D. M. Sladić, I. D. Brčeski and K. K. Anđelković, Inorg. Chem. Commun., 2006, 9, 862–865 CrossRef.
- T. R. Todorović, A. Bacchi, N. O. Juranić, D. M. Sladić, G. Pelizzi, T. T. Božić, N. R. Filipović and K. K. Anđelković, Polyhedron, 2007, 26, 3428–3436 CrossRef.
- S. Bjelogrlić, T. Todorović, A. Bacchi, M. Zec, D. Sladić, T. Srdić-Rajić, D. Radanović, S. Radulović, G. Pelizzi and K. Anđelković, J. Inorg. Biochem., 2010, 104, 673–682 CrossRef PubMed.
- N. Gligorijević, T. Todorović, S. Radulović, D. Sladić, N. Filipović, D. Gođevac, D. Jeremić and K. Anđelković, Eur. J. Med. Chem., 2009, 44, 1623–1629 CrossRef PubMed.
- T. Srdić-Rajić, M. Zec, T. Todorović, K. Anđelković and S. Radulović, Eur. J. Med. Chem., 2011, 46, 3734–3747 CrossRef PubMed.
- M. Zec, T. Srdić-Rajić, A. Krivokuća, R. Janković, T. Todorović, K. Andjelković and S. Radulović, Med. Chem., 2014, 10, 759–771 CrossRef CAS.
- M. Zec, T. Srdić-Rajić, A. Konić-Ristić, T. Todorović, K. Andjelković, I. Filipović-Ljesković and S. Radulović, Anti. Canc. Agents Med. Chem., 2014, 10, 1071–1080 Search PubMed.
- P. A. Sotiropoulou, M. S. Christodoulou, A. Silvani, C. Herold-Mende and D. Passarella, Drug Discovery Today, 2014, 19, 1547–1562 CrossRef CAS PubMed.
- J. P. Thiery, H. Acloque, R. Y. Huang and M. A. Nieto, Cell, 2009, 139, 871–890 CrossRef CAS PubMed.
- B. Beck and C. Blanpain, Nat. Rev. Cancer, 2013, 13, 727–738 CrossRef CAS PubMed.
- F. H. Sarkar, Y. Li, Z. Wang and D. Kong, Minerva Chir., 2009, 64, 489–500 CAS.
- T. Arumugam, V. Ramachandran, K. F. Fournier, H. Wang, L. Marquis and J. L. Abbruzzese, Cancer Res., 2009, 69, 5820–5828 CrossRef CAS PubMed.
- D. Kovala-Demertzi, P. N. Yadav, J. Wiecek, S. Skoulika, T. Varadinova and M. A. Demertzis, J. Inorg. Biochem., 2006, 100, 1558–1567 CrossRef CAS PubMed.
- D. Kovala-Demertzi, A. Alexandratos, A. Papageorgiou, P. N. Yadav, P. Dalezis and M. A. Demertzis, Polyhedron, 2008, 27, 2731–2738 CrossRef CAS.
- C. R. Kowol, E. Reisner, I. Chiorescu, V. B. Arion, M. Galanski, D. V. Deubel and B. K. Keppler, Inorg. Chem., 2008, 47, 11032–11047 CrossRef CAS PubMed.
- E. Lopez-Torres, M. A. Mendiola, J. Rodrıguez-Procopio, M. T. Sevilla, E. Colacio, J. M. Moreno and I. Sobrados, Inorg. Chim. Acta, 2001, 323, 130–138 CrossRef CAS.
- J. Wang, Analytical Electrochemistry, Wiley-VCH, New York, 2nd edn, 2000, p. 34 Search PubMed.
- P. S. Zhao, H. Y. Wang, J. Song and L. D. Lu, Struct. Chem., 2010, 21, 977–987 CrossRef CAS.
- A. Albert and E. P. Serjeant, The determination of Ionization constants, Chapman and Hall, London, 2nd edn, 1971, p. 44 Search PubMed.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, J. Appl. Crystallogr., 2008, 41, 466–470 CrossRef CAS.
- F. H. Allen, Acta Crystallogr., Sect. B: Struct. Sci., 2002, 58, 380–388 CrossRef.
- T. R. Todorović, A. Bacchi, D. M. Sladić, N. M. Todorović, T. T. Božić, D. D. Radanović, N. R. Filipović, G. Pelizzi and K. K. Anđelković, Inorg. Chim. Acta, 2009, 362, 3813–3820 CrossRef.
- E. Manoj and M. R. P. Kurup, Polyhedron, 2008, 27, 275–282 CrossRef CAS.
- X. Fan, J. Dong, R. Min, Y. Chen, X. Yi, J. Zhou and S. Zhang, J. Coord. Chem., 2013, 66, 4268–4279 CrossRef CAS.
- S. Zhang, J. Dong, X. Fan, Y. Chen and J. Zhou, J. Coord. Chem., 2012, 65, 3098–3110 CrossRef CAS.
- D. X. West, P. M. Ahrweiler, G. Ertem, J. P. Scovill, D. L. Klayman, J. L. Flippen-Anderson, R. Gilardi, C. George and L. K. Pannell, Transition Met. Chem., 1985, 10, 264–270 CrossRef CAS.
- M. T. Zimmerman, C. A. Bayse, R. R. Ramoutar and J. L. Brumaghim, J. Inorg. Biochem., 2015, 145, 30–40 CrossRef CAS PubMed.
- R. L. Prior, X. Wu and K. Schaich, J. Agric. Food Chem., 2005, 53, 4290–4302 CrossRef CAS PubMed.
- L. Galluzzi, I. Vitale, J. M. Abrams, E. S. Alnemri, E. H. Baehrecke, M. V. Blagosklonny, T. M. Dawson, V. L. Dawson, W. S. El-Deiry, S. Fulda, E. Gottlieb, D. R. Green, M. O. Hengartner, O. Kepp, R. A. Knight, S. Kumar, S. A. Lipton, X. Lu, F. Madeo, W. Malorni, P. Mehlen, G. Nuñez, M. E. Peter, M. Piacentini, D. C. Rubinsztein, Y. Shi, H. U. Simon, P. Vandenabeele, E. White, J. Yuan, B. Zhivotovsky, G. Melino and G. Kroemer, Cell Death Differ., 2012, 19, 107–120 CrossRef CAS PubMed.
- S. H. Lee, X. W. Meng, K. S. Flatten, D. A. Loegering and S. H. Kaufmann, Cell Death Differ., 2013, 20, 64–76 CrossRef CAS PubMed.
- D. V. Krysko, T. Vanden Berghe, K. D’Herde and P. Vandenabeele, Methods, 2008, 44, 205–221 CrossRef CAS PubMed.
- G. Kroemer and S. J. Martin, Nat. Med., 2005, 11, 725–730 CrossRef PubMed.
- D. R. McIlwain, T. Berger and T. W. Mak, Cold Spring Harbor Perspect. Biol., 2013, 5, a008656 Search PubMed.
- J. Puccini, L. Dorstyn and S. Kumar, Cell Death Differ., 2013, 20, 1133–1139 CrossRef CAS PubMed.
- A. Oleksi, A. G. Blanco, R. Boer, J. Usón, J. Aymamí, A. Rodger, M. J. Hannon and M. Coll, Angew. Chem., Int. Ed., 2006, 45, 1227–1231 CrossRef CAS PubMed.
- A. Terenzi, G. Barone, A. Silvestri, A. M. Giuliani, A. Ruggirello and V. T. Liveri, J. Inorg. Chem., 2009, 103, 1–9 CAS.
- R. Eshkourfu, B. Čobeljić, M. Vujčić, I. Turel, A. Pevec, K. Sepčić, M. Zec, S. Radulović, T. Srdić-Radić, D. Mitić, K. Andjelković and D. Sladić, J. Inorg. Biochem., 2011, 105, 1196–1203 CrossRef CAS PubMed.
- C. Y. Gao, X. Qiao, Z. Y. Ma, Z. G. Wang, J. Lu, J. L. Tian, J. Y. Xu and S. P. Yan, Dalton Trans., 2012, 41, 12220–12232 RSC.
- Z. Jannesari, H. Hadadzadeh, Z. Amirghofran, J. Simpson, T. Khayamian and B. Maleki, Spectrochim. Acta, Part A, 2015, 136, 1119–1133 CrossRef CAS PubMed.
- C. Icsel and V. T. Yilmaz, J. Photochem. Photobiol., B, 2014, 130, 115–121 CrossRef CAS PubMed.
- M. Cory, D. D. Mckee, J. Kagan, D. W. Henry and J. A. Miller, J. Am. Chem. Soc., 1985, 107, 2528–2536 CrossRef CAS.
- J. Wang, L. Shuai, X. Xiao, Y. Zeng, Z. Li and T. Matsumura-Inoue, J. Inorg. Biochem., 2005, 99, 883–885 CrossRef CAS PubMed.
- N. C. Garbett, N. B. Hammond and D. E. Graves, Biophys. J., 2004, 87, 3974–3981 CrossRef CAS PubMed.
- Y. Shi, C. Guo, Y. Sun, Z. Liu, F. Xu, Y. Zhang, Z. Wen and Z. Li, Biomacromolecules, 2011, 12, 797–803 CrossRef CAS PubMed.
- F. G. Loontiens, P. Regenfuss, A. Zechel, L. Dumortier and R. M. Clegg, Biochemistry, 1990, 29, 9029–9039 CrossRef CAS PubMed.
- A. Adhikary, V. Buschmann, C. Müller and M. Sauer, Nucleic Acids Res., 2003, 31, 2178–2186 CrossRef CAS PubMed.
- J. Chen and J. Stubbe, Curr. Opin. Chem. Biol., 2004, 8, 175–181 CrossRef CAS PubMed.
- B. Birdsall, R. W. King, M. R. Wheeler, C. A. Lewis, S.R. Goode, R. B. Dunlap and G. C. K. Roberts, Anal. Biochem., 1983, 132, 353–361 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer Science Business Media, New York, USA, 3rd edn, 2006, pp. 11, 56 and 289 Search PubMed.
- M. Amiri, K. Jankeje and J. R. Albani, J. Fluoresc., 2010, 20, 651–656 CrossRef CAS PubMed.
- M. R. Eftink and C. A. Ghiron, Anal. Biochem., 1981, 114, 199–227 CrossRef CAS PubMed.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef CAS PubMed.
- Y. Hu, Y. Liu, J. Wang, X. Xiao and S. Qu, J. Pharm. Biomed. Anal., 2004, 36, 915–919 CrossRef CAS PubMed.
- G. Sudlow, D. J. Birkett and D. N. Wade, Mol. Pharmacol., 1975, 11, 824–832 CAS.
- CrysAlisPro Software system, Agilent Technologies UK Ltd., Oxford, 2014 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- C. B. Hübschle, G. M. Sheldrick and B. Dittrich, J. Appl. Crystallogr., 2011, 44, 1281–1284 CrossRef PubMed.
- A. L. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 148–155 CrossRef CAS PubMed.
- S. Léonce, V. Pérez, S. Lambel, D. Peyroulan, F. Tillequin, S. Michel, M. Koch, B. Pfeiffer, G. Atassi, J. A. Hickman and A. Pierré, Mol. Pharmacol., 2001, 60, 1383–1391 CrossRef.
- M. E. Reichmann, S. A. Rice, C. A. Thomas and P. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef CAS.
- R. Vijayalakshmi, M. Kanthimathi, V. Subramanian and B. Unni Nair, Biochem. Biophys. Res. Commun., 2000, 271, 731–734 CrossRef CAS PubMed.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, USA, 2nd edn, 1989, pp. 133–134 Search PubMed.
- B. Ojha and G. Das, J. Phys. Chem. B, 2010, 114, 3979–3986 CrossRef CAS PubMed.
- F. L. Cui, J. Fan, J. P. Li and Z. D. Hu, Bioorg. Med. Chem., 2004, 12, 151–157 CrossRef CAS PubMed.
- S. Grimme, J. Comput. Chem., 2006, 27, 1787–1799 CrossRef CAS PubMed.
- A. Schaefer, H. Horn and R. Ahlrichs, J. Chem. Phys., 1992, 97, 2571–2577 CrossRef CAS.
- A. Schaefer, C. Huber and R. Ahlrichs, J. Chem. Phys., 1994, 100, 5829–5835 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 (Revision D.01), Gaussian Inc., Wallingford CT, 2013 Search PubMed.
- R. Dennington, T. Keith and J. Millam, GaussView, version 5, Semichem Inc., Shawnee Mission, KS, 2009 Search PubMed.
- D. G. Wei, W. D. Wilson and S. Neidle, J. Am. Chem. Soc., 2013, 135, 1369–1377 CrossRef CAS PubMed.
- W. Zhu, Y. Wang, K. Li, J. Gao, C. H. Huang, C. C. Chen, T. P. Ko, Y. Zhang, R. T. Guo and E. Oldfield, J. Med. Chem., 2015, 58, 1215–1227 CrossRef CAS PubMed.
- S. Curry, H. Mandelkow, P. Brick and N. Franks, Nat. Struct. Biol., 1998, 5, 827–835 CrossRef CAS PubMed.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38–52 CrossRef CAS PubMed.
- Z. M. Wang, J. X. Ho, J. R. Ruble, J. Rose, F. Ruker, M. Ellenburg, R. Murphy, J. Click, E. Soistman, L. Wilkerson and D. C. Carter, Biochim. Biophys. Acta, 2013, 1830, 5356–5374 CrossRef CAS PubMed.
- M. F. Sanner, J. Mol. Graphics Modell., 1999, 17, 57–61 CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 16, 2785–2791 CrossRef PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
Footnote |
† Electronic supplementary information (ESI) available: FT-IR (Fig. S1) and NMR (Fig. S2–S9) spectra; cyclic voltammograms (Fig. S10); pH dependent UV-Vis spectra (Fig. S11 and S12); packing diagrams (Fig. S13 and S14); sigmoidal dose–response curves (Fig. S15); Stern–Volmer plot (Fig. S16); modified Stern–Volmer plot (Fig. S17); the plot of ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka vs. 1/T (Fig. S18); double-log plot (Fig. S19); fluorescence spectral overlap (Fig. S20); conformations of the complex in the DNA duplex (Fig. S21); the location of the HSA–Zn complex binding site (Fig. S22); numbering of atoms (Scheme S1); hydrogen bonding geometry (Table S1); interactions of HSA binding site (Table S2). CCDC 1413656 and 1413657. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5ra19849f Ka vs. 1/T (Fig. S18); double-log plot (Fig. S19); fluorescence spectral overlap (Fig. S20); conformations of the complex in the DNA duplex (Fig. S21); the location of the HSA–Zn complex binding site (Fig. S22); numbering of atoms (Scheme S1); hydrogen bonding geometry (Table S1); interactions of HSA binding site (Table S2). CCDC 1413656 and 1413657. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5ra19849f |
| This journal is © The Royal Society of Chemistry 2015 |