Open Access Article
Open Access ArticlePolymerization-based signal amplification for paper-based immunoassays†
Abraham K.
Badu-Tawiah‡
a,
Shefali
Lathwal‡
b,
Kaja
Kaastrup
b,
Mohammad
Al-Sayah
ac,
Dionysios C.
Christodouleas
a,
Barbara S.
Smith
a,
George M.
Whitesides
*a and
Hadley D.
Sikes
*b
aDepartment of Chemistry and Chemical Biology, Harvard University, 12 Oxford Street, Cambridge, MA 02138, USA. E-mail: gwhitesides@gmwgroup.harvard.edu
bDepartment of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. E-mail: sikes@mit.edu
cDepartment of Biology, Chemistry and Environmental Sciences, American University of Sharjah, P.O. Box 26666, Sharjah, United Arab Emirates
First published on 18th November 2014
Abstract
Diagnostic tests in resource-limited settings require technologies that are affordable and easy to use with minimal infrastructure. Colorimetric detection methods that produce results that are readable by eye, without reliance on specialized and expensive equipment, have great utility in these settings. We report a colorimetric method that integrates a paper-based immunoassay with a rapid, visible-light-induced polymerization to provide high visual contrast between a positive and a negative result. Using Plasmodium falciparum histidine-rich protein 2 as an example, we demonstrate that this method allows visual detection of proteins in complex matrices such as human serum and provides quantitative information regarding analyte levels when combined with cellphone-based imaging. It also allows the user to decouple the capture of analyte from signal amplification and visualization steps.
Introduction
Analytical devices made of cellulosic materials (e.g. filter and chromatography paper) are attractive for resource-limited settings (RLS) because of their low cost,1–3 ease of fabrication,3 and porous structure that facilitates capillary flow. 2D and 3D microfluidic paper-based analytical devices (μPADs) have been developed and used for detection of small molecules,2 metals,4 and proteins.1,2,5 A crucial component of an analytical test is the mechanism that allows users to read out the results of a test. A wide range of visualization mechanisms are available for use with paper surfaces,6 but POC tests in RLS are constrained by cost and limited infrastructure. Colorimetric methods based on enzymatic reactions or gold nanoparticles (AuNP) are, therefore, widely used in paper-based POC tests because they provide an equipment-free readout.6 These methods sometimes produce low visual contrast between a positive and a negative result5,7 that leads to subjectivity in interpretation of the result by the user, along with decreases in accuracy and sensitivity. Color development after capture of the analyte from a patient sample by a bioactive cellulose surface can require intervals from 30 minutes for enzymatic reactions8 to 20–30 minutes for AuNP-based sensing with silver enhancement5 and can give a false negative result if read prematurely. These tests can also give a false positive result if read a few minutes after a specified end time.9 The motivation for this work was to develop a polymerization-based colorimetric sensing method for use with paper immunoassays to address these limitations of existing colorimetric methods.A polymerization response can be coupled to detection of an analyte using a method termed polymerization-based amplification (PBA).10–13 In PBA, photoinitiators are localized to regions where specific molecular binding events have occurred through covalent coupling of the photoinitiator to one of the affinity reagents used in the assay. When the photoinitiator molecules are supplied with an appropriate dose of light in the presence of acrylate monomers, they initiate a free-radical polymerization reaction to generate an interfacial hydrogel. The result of the polymerization process is the formation of a hydrogel only in areas where the local concentration of the photoinitiator near a binding surface is sufficient to overcome competing inhibition reactions, and to initiate polymerization. PBA is rapid with a reaction time of less than 100 seconds and can be performed in air, without the need for oxygen removal via purging,13 by using an eosin/tertiary amine initiation system that can overcome oxygen inhibition through an eosin regeneration mechanism.14 The reactants and key elementary reactions in the overall polymerization reaction are described in the ESI.† PBA was previously developed using bioactive glass surfaces and the colorless hydrogel was swollen with a dye solution to aid visualization by eye.12,13 However, the swelling method could not be used with paper because dyes adhere non-specifically to paper. This non-specific adhesion led to low contrast between the hydrogel and the background (ESI†). In this work, we have adapted PBA to detect molecular binding events on a paper surface. We developed a new visualization method by using the pH-dependent color change of a pH indicator to detect the formation of the hydrogel. We have successfully used this polymerization method to generate colorimetric results, easily perceptible to the unaided eye, for the immuno-detection of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) on modified chromatography paper. PfHRP2 is a soluble protein released into the blood stream during infection by Plasmodium falciparum. It is a well-established biomarker for falciparum infection and several commercial diagnostic tests are based on its detection.15
Method development
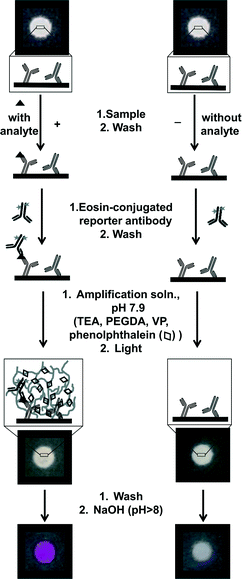
A monoclonal capture antibody (Arista Biologicals, Clone 44) was covalently coupled to the aldehyde groups (ESI,† Fig. S1) in a hydrophilic test zone of oxidized chromatography paper (ESI,† Fig. S2). When this test zone was contacted with a solution containing PfHRP2, the capture antibody formed a complex with PfHRP2 (Fig. 1). In order to associate a polymerization response with the presence of PfHRP2 on the surface, we coupled eosin, a photoinitiator, to a monoclonal reporter antibody (Arista Biologicals, Clone 45) (ESI,† Fig. S3). When a paper surface containing the capture antibody-PfHRP2 complex was contacted with the eosin-modified reporter antibody, the reporter bound to PfHRP2. This binding step localized eosin where PfHRP2 had been captured, resulting in a positive test surface. In contrast, for a negative test surface, eosin was not localized if PfHRP2 was not present to bridge the two antibodies. We placed positive and negative test surfaces prepared in this way in contact with a 20 μL drop of an aqueous amplification solution and illuminated them from above using an array of light-emitting diodes (LEDs) (λ = 522 nm, 30 mW cm−2). The amplification solution contained 150 mM triethanolamine (TEA), 200 mM (poly)ethyleneglycol diacrylate (PEGDA), 100 mM 1-vinyl-2-pyrrolidinone (VP), 1.6 mM phenolphthalein, and 0.35 μM free eosin. The pH of the solution was adjusted to 7.9 using hydrochloric acid so that phenolphthalein was present in its colorless form (ESI,† Fig. S4) and did not compete with eosin for absorption of light (ESI,† Fig. S5). The illumination time was chosen such that the photoinitiator density was sufficient to overcome oxygen inhibition and initiate polymerization on the positive test surfaces, but not on the negative test surfaces. As the hydrogel formed, phenolphthalein that was present in the aqueous solution became physically trapped in the transparent cross-linked network. The hydrogel remained on paper even after the surface was washed to remove unreacted amplification solution. When 2 μL of 0.5 M NaOH was added to the surface, the pH increased above 8 and phenolphthalein changed from colorless to bright pink. The change in color occurred immediately upon the addition of NaOH (ESI†) and allowed visual detection of the hydrogel by the unaided eye. In contrast, no hydrogel formed on the negative test surfaces and no change in color was observed upon addition of NaOH to these surfaces. The pH-dependent hydrogel visualization thus allowed us to differentiate clearly between a positive and a negative test surface based on a colorimetric response (Fig. 1). Each component of the amplification solution used to detect molecular recognition at a paper surface served a unique purpose that is further discussed in the ESI.†Results and discussion
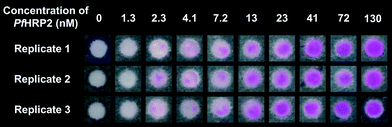
We optimized the paper-based immunoassay (ESI,† Fig. S6–S9) and determined the limit-of-detection (LoD) of PfHRP2 using PBA by testing nine different concentrations of PfHRP2 in buffer ranging over two orders of magnitude, 1.3 nM to 130 nM. The negative surfaces were contacted with a buffered solution without PfHRP2. All surfaces were illuminated for 90 seconds and the results were imaged using a cellphone. The average intensity of the colorimetric response was quantified using ImageJ (ESI,† Table S1–S2). We used two approaches to define LoD, i) visual LoD, the concentration where all the replicates show a visible colorimetric response, and ii) calculated LoD, the minimum concentration that gives an average colorimetric intensity that is higher than the average intensity from the negative surface by three times the standard deviation of the mean from the negative surface. Representative images from one of five independent dose–response trials are shown in Fig. 2A. The visual LoD was 7.2 nM because all replicates (n = 19) with a PfHRP2 concentration of 7.2 nM and higher showed a bright pink hydrogel. All replicates with PfHRP2 concentrations of 2.3 nM, 1.3 nM, or 0 nM (negative surfaces) did not have a visible hydrogel. At a PfHRP2 concentration of 4.1 nM, 9 out of a total of 19 replicates showed a visible hydrogel. This response is consistent with the threshold nature of PBA10 just below the limit of detection, where small differences in the number of bound proteins can push the initiator density either slightly above or slightly below the concentration threshold where propagation reactions become competitive with inhibition reactions.16 The calculated LoD (Fig. 2B) was 5.8 nM according to the definition given above. The close agreement between the visual and the calculated LoD indicates that if a hydrogel is present on the surface, it can be reliably detected by unaided eye. Therefore, for qualitative assays where the exact concentration of analyte is not needed for making clinical decisions, PBA can be a useful tool for unambiguous visual detection. At the same time, if quantitative information about an analyte is required, paper-based PBA can be used in conjunction with cellphone-based image capture (ESI†).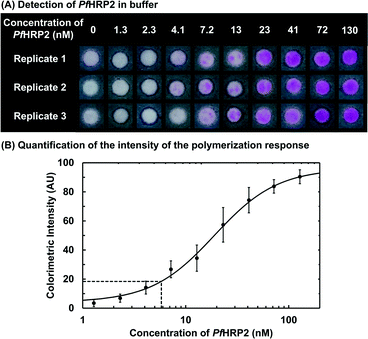 | ||
| Fig. 2 Detection of PfHRP2 (0–130 nM) in a buffered solution by using polymerization-based amplification on paper. (A) Representative images of the colorimetric results for detection of various concentrations of PfHRP2. The visual LoD, the minimum concentration at which all replicates give a positive colorimetric response, was 7.2 nM (0.56 μg mL−1). (B) Quantitation of the intensity of the colorimetric results for detection of PfHRP2. The data were fitted to a sigmoidal curve (ESI†). Each data point is an average of eight replicates and error bars indicate standard deviation. The vertical line indicates the calculated LoD of PfHRP2, 5.8 nM and the horizontal line shows the corresponding intensity. | ||
We evaluated the effect of a complex matrix such as serum on the performance of the assay. In brief, we tested nine different concentrations, 130 nM to 1.3 nM, of PfHRP2 prepared in human serum. Undiluted human serum, without any added PfHRP2, was used for the negative surfaces. An illumination time of 50 seconds (ESI†) was used to differentiate between positive and negative responses. Fig. 3 shows that the visual LoD of PfHRP2 in human serum is similar to the visual LoD obtained in buffer. The mean concentration of plasma PfHRP2 has been reported as 28 nM in a cohort of 337 adult patients with falciparum malaria.17 This concentration falls well within the dynamic range of the polymerization-based colorimetric method that we have developed.
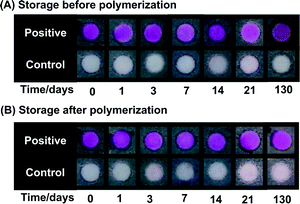
We further assessed the robustness of the assay by testing the stability of the photoinitiator and the stability of the phenolphthalein trapped in the hydrogel. We performed two different storage experiments. In a first experiment, positive and negative test surfaces were stored in air, at room temperature in a closed drawer for 1, 3, 7, 14, 21, and 130 days. After the specified duration, the surfaces were contacted with the aqueous amplification solution, irradiated for 90 seconds and visualized using NaOH. The similarity in the colorimetric readout (Fig. 4A) indicates that eosin is stable on the surface and can initiate the polymerization reaction after storage for extended periods of time. In a second experiment, positive and negative test surfaces were prepared, contacted with amplification solution and illuminated for 90 seconds on the same day. The surfaces were washed to remove the unreacted solution, and stored in a drawer for 1, 3, 7, 14, 21 and 130 days before NaOH was added for visualization. Fig. 4B shows that the hydrogel resulting from the polymerization process can be rehydrated with NaOH solution after up to four months in storage to generate a strong colorimetric response, indicating that phenolphthalein remains trapped and functional in the hydrogel.
The above storage experiments demonstrate that using PBA with paper surfaces provides two separate points where the assay can be stopped, stored and restarted. This ability adds flexibility to the assay and can also eliminate sample collection and storage because it creates the possibility of self-testing by patients at home followed by polymerization and diagnosis later at a health facility.
Conclusions
We have described a polymerization-based sensing method with a pH-responsive colorimetric readout for use with paper immunoassays that: i) is rapid; it takes less than 100 seconds for the polymerization reaction after capture of analyte and color development is instantaneous upon addition of NaOH ii) generates results that are easily perceptible using the unaided eye, iii) provides the flexibility to store the assay and control the start of the polymerization and the visualization steps, and iv) provides quantitative information when combined with cellphone-based image capture and image processing. We have used a sandwich immunoassay for PfHRP2 detection as a model, but this method can be easily adapted for all types of immunoassays (direct or indirect). The advantages of PBA on paper come at the cost of requiring a source of light for illumination. The choice of an appropriate illumination time is necessary for PBA to differentiate between a positive and a negative test. However, for a fixed light source, the illumination time for a given type of sample needs to be chosen only once and is reproducible (ESI†). We used a portable, electricity-powered LED light source in the laboratory, but we can envision a portable, battery-powered LED device that can be adapted for use in RLS.In comparison to colorimetric methods that are currently used with paper-based immunoassays, PBA requires two additional assay steps but ten-fold less time for the signal amplification and visualization steps – 2–2.5 minutes compared to 20–30 minutes for enzymatic and AuNP-based methods (ESI,† Table S3). This reduction in time combined with the ability to stop, store and restart the PBA-based assay has the potential to minimize false readouts due to time constraints in situations where only a few health workers are tending to the needs of many patients. The high visual contrast provided by the PBA system, even close to the LoD, also makes it easier for a user to visually interpret the results, in comparison to enzymatic and AuNP-based methods where low contrast (e.g.ref. 5) can lead to ambiguous visual interpretation.
Because the focus of this work was on development of the polymerization method, rather than building a complete device, we used a flow-through system where a user performs wash steps in the immunoassay and adds the next solution. Strategies that have been used successfully to reduce the number of steps performed by users, such as implementation of assays in a 2D paper network18 or a paper-based 3D microfluidic format,19 could be used with paper-based PBA assays as well. Although the new method is not yet ready for field-testing, we expect that the advantages provided by PBA-based colorimetric sensing hold promise for reducing inaccurate reading of results in rapid screening tests in field settings.
Acknowledgements
The Bill and Melinda Gates Foundation (award no. 51308) and the Defense Advanced Research Projects Agency (HR0011-12-2-0010) supported this work. The opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. M.A. thanks the Arab Fund Fellowship Program (2-1-72/1841) and his institution for a sabbatical leave award. We acknowledge a Landau Chemical Engineering Practice School Fellowship (SL), an NSF Graduate Research Fellowship (KK), and a Burroughs Wellcome Fund Career Award at the Scientific Interface (HDS).Notes and references
- N. R. Pollock, J. P. Rolland, S. Kumar, P. D. Beattie, S. Jain, F. Noubary, V. L. Wong, R. A. Pohlmann, U. S. Ryan and G. M. Whitesides, Sci. Transl. Med., 2012, 4, 152ra129 CrossRef.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., Int. Ed., 2007, 46, 1318–1320 CrossRef CAS.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS.
- M. M. Mentele, J. Cunningham, K. Koehler, J. Volckens and C. S. Henry, Anal. Chem., 2012, 84, 4474–4480 CrossRef CAS.
- R. C. Murdock, L. Shen, D. K. Griffin, N. Kelley-Loughnane, I. Papautsky and J. A. Hagen, Anal. Chem., 2013, 85, 11634–11642 CrossRef CAS.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- K. Abe, K. Kotera, K. Suzuki and D. Citterio, Anal. Bioanal. Chem., 2010, 398, 885–893 CrossRef CAS.
- C.-M. Cheng, A. W. Martinez, J. Gong, C. R. Mace, S. T. Phillips, E. Carrilho, K. A. Mirica and G. M. Whitesides, Angew. Chem., 2010, 122, 4881–4884 CrossRef.
- C. D. Chin, T. Laksanasopin, Y. K. Cheung, D. Steinmiller, V. Linder, H. Parsa, J. Wang, H. Moore, R. Rouse, G. Umviligihozo, E. Karita, L. Mwambarangwe, S. L. Braunstein, J. Van De Wijgert, R. Sahabo, J. E. Justman, W. El-sadr and S. K. Sia, Nat. Med., 2011, 17, 1015–1019 CrossRef CAS.
- H. D. Sikes, R. R. Hansen, L. M. Johnson, R. Jenison, J. W. Birks, K. L. Rowlen and C. N. Bowman, Nat. Mater., 2008, 7, 52–56 CrossRef CAS.
- R. R. Hansen, H. D. Sikes and C. N. Bowman, Biomacromolecules, 2008, 9, 355–362 CrossRef CAS.
- L. R. Kuck and A. W. Taylor, BioTechniques, 2008, 45, 179–186 CrossRef CAS.
- K. Kaastrup and H. D. Sikes, Lab Chip, 2012, 12, 4055–4058 RSC.
- H. J. Avens and C. N. Bowman, J. Polym. Sci., Part A: Polym. Chem., 2010, 47, 6083–6094 CrossRef.
- World Health Organization, Malaria Rapid Diagnostic Test Performance - Results of WHO product testing of malaria RDTs: Round 4 (2012), 2012, vol. 4 Search PubMed.
- K. Kaastrup, L. Chan and H. D. Sikes, Anal. Chem., 2013, 85, 8060 CrossRef.
- A. M. Dondorp, V. Desakorn, W. Pongtavornpinyo, D. Sahassananda, K. Silamut, K. Chotivanich, P. N. Newton, P. Pitisuttithum, A. M. Smithyman, N. J. White and N. P. J. Day, PLoS Med., 2005, 2, e204 Search PubMed.
- E. Fu, T. Liang, P. Spicar-Mihalic, J. Houghtaling, S. Ramachandran and P. Yager, Anal. Chem., 2012, 84, 4574–4579 CrossRef CAS.
- L. Ge, S. Wang, X. Song, S. Ge and J. Yu, Lab Chip, 2012, 12, 3150–3158 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: See DOI: 10.1039/c4lc01239a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |