Open Access Article
Open Access ArticleThe dissolution and biological effects of silver nanoparticles in biological media
K.
Loza
a,
J.
Diendorf
a,
C.
Sengstock
b,
L.
Ruiz-Gonzalez
c,
J. M.
Gonzalez-Calbet
c,
M.
Vallet-Regi
c,
M.
Köller
b and
M.
Epple
*a
aInorganic Chemistry and Centre for Nanointegration Duisburg-Essen (CeNIDE), University of Duisburg-Essen, Universitaetsstr. 5-7, 45117 Essen, Germany. E-mail: matthias.epple@uni-due.de
bBergmannsheil University Hospital/Surgical Research, Ruhr-University of Bochum, Buerkle-de-la-Camp-Platz 1, 44789 Bochum, Germany
cDepartmento de Quimica Inorganica, Universidad Complutense, 28040 Madrid, Spain
First published on 10th January 2014
Abstract
Silver ions and silver nanoparticles have a well-known biological effect that typically occurs in biological or environmental media of complex composition. Silver nanoparticles release silver ions if oxidizing species like molecular oxygen or hydrogen peroxide are present. The presence of glucose as a model for reducing sugars has only a small effect on the dissolution rate. In the presence of chloride ions, precipitation of silver chloride nanoparticles occurs. At physiological salt concentrations, no precipitation of silver phosphate occurs as the precipitation of silver chloride always occurs first. If the surface of a silver nanoparticle is passivated by cysteine, the dissolution is quantitatively inhibited. Upon immersion of silver nanoparticles in pure water for 8 months, leading to about 50% dissolution, no change in the surface was observed by transmission electron microscopy. A model for the dissolution was derived from immersion and dissolution experiments in different media and from high-resolution transmission electron microscopy. A literature survey on the available data on the dissolution of silver nanoparticles showed that only qualitative trends can be identified as the nature of the nanoparticles and of the immersion medium are practically never comparable. The dissolution effects were confirmed by cell culture experiments (human mesenchymal stem cells and neutrophil granulocytes) where silver nanoparticles that were stored under argon had a clearly lower cytotoxicity than those stored under air. They also led to a less formation of reactive oxygen species (ROS). This underscores that silver ions are the toxic species.
Introduction
Silver nanoparticles are widely used in consumer products and in medicine due to their antibacterial action.1–10 However, there is increasing evidence that the release of silver ions from nanoparticles is responsible for this biological effect. Xiu et al. showed in a very elegant experiment that silver nanoparticles did not show a bactericidal effect if the bacteria were cultivated under anaerobic conditions, i.e. when no dissolution of the nanoparticles occurred.11 Studies on the dissolution behaviour were reported in a number of publications,9,11–22 but there is no agreement on the dissolution behaviour in more complex media which are present, e.g. in biological systems and in the environment.Based on our previous results on the dissolution of dispersed silver nanoparticles in pure water,15 we have extended our studies to more complex media which are closer to the biological conditions in cell culture media and body fluids. We have analysed the dissolution kinetics and the nature of the silver nanoparticles after immersion in different media for up to 4000 h. Table 1 summarizes the results of dissolution experiments from the literature and from this study.
| Functionalization of silver nanoparticles | Diameter/nm | Zeta potential/mV | Silver concentration/g L−1 | Temperature/°C | Immersion medium | Immersion time/h | Degree of dissolution/% | Ref. |
|---|---|---|---|---|---|---|---|---|
| Citrate | 85 | −30 | 0.32 | 25 | Water | 336 | 15 | 15 |
| Citrate | 85 | −30 | 0.14 | 25 | Water | 410 | 12 | 15 |
| Citrate | 85 | −30 | 0.32 | 37 | Water | 307 | 56 | 15 |
| Citrate | 85 | −30 | 0.14 | 37 | Water | 444 | 53 | 15 |
| Citrate | 4.8 | −45 | 0.002 | 25 | Air-saturated distilled water, [O2] = 9.1 mg L−1; pH = 5.68 | 3000 | ∼100 | 12 |
| Citrate | 4.8 | −45 | 0.0002 | 25 | Air-saturated distilled water, [O2] = 9.1 mg L−1; pH = 5.68 | 240 | ∼100 | 12 |
| Citrate | 4.8 | −45 | 0.00005 | 25 | Air-saturated distilled water, [O2] = 9.1 mg L−1; pH = 5.68 | 120 | ∼100 | 12 |
| Citrate | 4.8 | −45 | 0.00005 | 4 | Air-saturated distilled water, [O2] = 9.1 mg L−1; pH = 5.68 | 120 | 36 | 12 |
| Citrate | 4.8 | −45 | 0.00005 | 37 | Air-saturated distilled water, [O2] = 9.1 mg L−1; pH = 5.68 | 120 | 96 | 12 |
| PVP | 50 | −17 | 0.22 | 5 | Water | 411 | 9 | 15 |
| PVP | 50 | −17 | 0.1 | 5 | Water | 149 | 8 | 15 |
| PVP | 50 | −17 | 0.05 | 5 | Water | 149 | 12 | 15 |
| PVP | 50 | −17 | 0.35 | 25 | Water | 134 | 48 | 15 |
| PVP | 50 | −17 | 0.1 | 25 | Water | 81 | 52 | 15 |
| PVP | 50 | −17 | 0.05 | 25 | Water | 42 | 43 | 15 |
| PVP | 50 | −17 | 0.35 | 37 | Water | 27 | 52 | 15 |
| PVP | 50 | −17 | 0.1 | 37 | Water | 27 | 89 | 15 |
| PVP | 50 | −17 | 0.05 | 37 | Water | 11 | 68 | 15 |
| PVP | 70 | −22 | 0.35 | 25 | Oxygen-free water | 1660 | 2 | This work |
| Citrate | 4.8 | −45 | 0.00005 | 25 | Natural seawater, pH = 7.90 | 192 | 38 | 12 |
| Citrate | 20 | — | 0.0003 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 40 | 21 |
| Citrate | 20 | — | 0.0006 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 29 | 21 |
| Citrate | 40 | — | 0.0003 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 21 | 21 |
| Citrate | 40 | — | 0.0006 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 20 | 21 |
| Citrate | 80 | — | 0.0003 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 10 | 21 |
| Citrate | 80 | — | 0.0006 | 25 | Modified ¼ Hoagland medium, [O2] = 7.8 mg L−1; pH = 5.6 | 336 | 8 | 21 |
| PVP | 70 | −22 | 0.35 | 25 | 0.9% NaCl | 3670 | 8 | This work |
| PVP | 70 | −22 | 0.35 | 25 | PBS | 3670 | 4 | This work |
| PVP | 39 | Negative | 1 | 25 | 0.01 M NaNO3, pH = 7 | 720 | 1.9 | 18 |
| PVP, sulphidated | 39 | Negative | 1 | 25 | 0.01 M NaNO3, pH = 7 | 730 | ca. 0 to 0.27 | 18 |
| PVP | 70 | −22 | 0.35 | 25 | 10 mM H2O2 | 1821 | 90 | This work |
| Citrate | 4.8 | −45 | 0.002 | 25 | Acetate buffer, pH 5.6 | 24 | 15 | 13 |
| Citrate | 4.8 | −45 | 0.002 | 25 | Acetate buffer, 0.4 mM citrate; pH 5.6 | 24 | 9 | 13 |
| Citrate | 4.8 | −45 | 0.002 | 25 | Acetate buffer, 10 mM citrate; pH 5.6 | 24 | 7 | 13 |
| Citrate | 4.8 | −45 | 0.002 | 25 | Acetate buffer, 0.4 mM Na2S; pH 5.6 | 24 | <1 | 13 |
| Citrate | 4.8 | −45 | 0.002 | 25 | Acetate buffer, 4 mM 11-mercaptoundecanoic acid; pH 5.6 | 24 | ca. 0 | 13 |
| PVP | 70 | −22 | 0.35 | 25 | 1 g L−1 cysteine | 4366 | ca. 0 | This work |
| PVP | 70 | −22 | 0.35 | 25 | 1 g L−1 glucose | 4366 | 61 | This work |
| Citrate | 4.8 | −45 | 0.00005 | 25 | Boric acid and bicarbonate low ionic strength seawater buffer, pH = 7.90 | 192 | 54 | 12 |
| Citrate | 23 | — | 0.005 | 25 | DMEM | 22 | ca. 75 | 20 |
| Citrate | 23 | — | 0.1 | 25 | DMEM | 22 | ca. 7 | 20 |
| PVP | 23 | — | 0.005 | 25 | DMEM | 22 | ca. 76 | 20 |
| 5 kDa PEG | 23 | — | 0.005 | 25 | DMEM | 22 | ca. 83 | 20 |
| 20 kDa PEG | 23 | — | 0.005 | 25 | DMEM | 22 | ca. 75 | 20 |
| Dextran | 23 | — | 0.005 | 25 | DMEM | 22 | ca. 67 | 20 |
| 5 kDa PEG | 23 | — | 0.1 | 25 | DMEM | 22 | ca. 11 | 20 |
| 20 kDa PEG | 23 | — | 0.1 | 25 | DMEM | 22 | ca. 9 | 20 |
| PVP | 4–5 | Negative | 0.005 | 37 | Synthetic gastric acid, pH = 1.12 | 24 | ∼98 | 23 |
| PVP | 4–5 | Negative | 0.005 | 37 | Pseudoextracellular fluid, pH = 7.52 | 24 | ∼80 | 23 |
It must be stressed that it is not possible to monitor the dissolution of nanoparticles and their cytotoxicity in the same experiment. The nanoparticles will dissolve with a given kinetics, and the cells will react with some delay to the increasing silver concentration. Therefore, we have separated these two aspects by measuring the dissolution kinetics of the silver nanoparticles in more complex media and by studying the cytotoxicity of silver nanoparticles that were stored under argon (i.e. without released silver ions up to the start of the experiment). Summarizing these observations, we propose a model for the behaviour of silver nanoparticles and for the state of released silver ions under these conditions.
Experimental section
Synthesis of silver nanoparticles
PVP-coated silver nanoparticles were synthesized by reduction with glucose in the presence of PVP according to Wang et al.24 Briefly, 2 g glucose and 1 g PVP were dissolved in 40 g water and heated to 90 °C. Then 0.5 g AgNO3 dissolved in 1 mL water was quickly added. The dispersion was kept at 90 °C for 1 h and then let to cool to room temperature. The particles were collected by ultracentrifugation (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g; 30 min), redispersed in pure water and collected again by ultracentrifugation. Thereby NO3−, excess glucose and its oxidation products, excess PVP, and excess Ag+ were all removed. The silver nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
400g; 30 min), redispersed in pure water and collected again by ultracentrifugation. Thereby NO3−, excess glucose and its oxidation products, excess PVP, and excess Ag+ were all removed. The silver nanoparticles were then redispersed in water by ultrasonication. The final silver concentration in all dispersions was determined by atomic absorption spectroscopy (AAS).
Poly(vinylpyrrolidone) (PVP K30, Povidon 30; Fluka, molecular weight 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), silver nitrate (Roth, p.a.), sodium chloride (VWR, p.a.), D-(+)-glucose (Fluka, p.a.), cysteine (Merck, p.a.), and hydrogen peroxide (Sigma, p.a.) were used. Phosphate-buffered saline solution (PBS) was obtained from Gibco. RPMI 1640 was obtained from Gibco. LB medium was obtained from Sigma-Aldrich. Fetal calf serum (FCS) was obtained from Gibco. Ultrapure water was prepared with an ELGA Purelab ultra instrument.
000 g mol−1), silver nitrate (Roth, p.a.), sodium chloride (VWR, p.a.), D-(+)-glucose (Fluka, p.a.), cysteine (Merck, p.a.), and hydrogen peroxide (Sigma, p.a.) were used. Phosphate-buffered saline solution (PBS) was obtained from Gibco. RPMI 1640 was obtained from Gibco. LB medium was obtained from Sigma-Aldrich. Fetal calf serum (FCS) was obtained from Gibco. Ultrapure water was prepared with an ELGA Purelab ultra instrument.
Analytical methods
Scanning electron microscopy (SEM) was performed with a FEI Quanta 400 ESEM instrument in a high vacuum after sputtering with Au:Pd, if necessary. Energy-dispersive X-ray spectrometry was carried out with a Genesis 4000 instrument. Transmission electron microscopy (TEM) was carried out both with JEOL 400EX and JEOL 3000FEG electron microscopes, equipped with an Oxford LINK EDS analyser. The samples were ultrasonically dispersed in ethanol and then transferred to holey carbon-coated copper grids.The hydrodynamic diameter and the zeta potential of the nanoparticles were measured by dynamic light scattering (DLS) using a Malvern Zetasizer Nano ZS. The polydispersity index (PDI) was below 0.3 in all cases. The concentration of silver was determined by atomic absorption spectroscopy (AAS; Thermo Electron Corporation, M-Series). The detection limit was 1 μg L−1.
X-ray powder diffraction was carried out on a Bruker D8 Advance instrument in Bragg-Brentano mode with Cu Kα radiation (1.54 Å). Dissolved oxygen was measured with a WTW Oxi3250 instrument equipped with a CellOx 325 sensor.
Dialysis experiments were carried out in dialysis bags (Spectra/Por Biotech; cellulose ester; MWCO 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) filled with 4 mL silver nanoparticle dispersion (0.35 g L−1 Ag) and immersed in 396 mL ultrapure water. The dialysis was carried out under slow stirring with a magnetic stirrer at room temperature.
000) filled with 4 mL silver nanoparticle dispersion (0.35 g L−1 Ag) and immersed in 396 mL ultrapure water. The dialysis was carried out under slow stirring with a magnetic stirrer at room temperature.
The dispersions of silver nanoparticles in different media were stirred at room temperature for 7 days. The nanoparticles and all precipitates were isolated by ultracentrifugation (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900g; 30 min), redispersed in pure water and again subjected to ultracentrifugation. This procedure was repeated three times.
900g; 30 min), redispersed in pure water and again subjected to ultracentrifugation. This procedure was repeated three times.
Simulation of the equilibrium of aqueous silver in the biological media was performed using Visual MINTEQ 3.0 (J.P. Gustafsson, Stockholm, 2011). The simulation was performed for 0.1 g L−1 dissolved silver. The model medium contained only inorganic salts. Solids were allowed to precipitate.
Preparation of stock and working solutions
PVP-coated spherical silver nanoparticles were dispersed in sterile ultrapure water at 1 g L−1 (1000 ppm) as stock solution. A solution of silver acetate with 1 g L−1 was the second stock solution. Both stock solutions were diluted with ultrapure water to prepare working stock solutions for the individual experiments at different silver concentrations (10–1000 ppm). The final concentrations were achieved by the addition of 50 μL of either silver nanoparticle or silver acetate working stock solutions to a 1 mL cell suspension prior to cultivation. Thereby the concentrations of other medium ingredients were kept constant in all experiments.The final concentrations used for bacterial and cell culture tests were 50 μg mL−1, 30 μg mL−1, 25 μg mL−1, 20 μg mL−1, and 12.5 μg mL−1 for silver nanoparticles. The nanoparticles were freshly prepared and stored for 1 month at 7 °C before the biological tests. Ag-NPs were stored either under air or under argon to avoid oxidation and subsequent release of silver ions.12,13,15,16,25
All concentration values given in tables and figures refer to the mass of silver in the solution, regardless of its state (ions or nanoparticles).
Cell culture experiments
Human mesenchymal stem cells (hMSCs, 3rd to 7th passage, Lonza, Walkersville Inc., MD, USA) were cultured in cell culture medium RPMI/10% FCS using 24-well cell culture plates (Falcon, Becton Dickinson GmbH, Heidelberg, Germany). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. hMSCs were sub-cultivated every 7–14 days depending on the cell proliferation. Adherent cells were washed with phosphate-buffered saline solution (PBS, GIBCO, Life Technologies) and detached from the culture flasks by the addition of 0.2 mL cm−2 0.25% trypsin/0.1% ethylenediaminetetraacetic acid (EDTA, Sigma-Aldrich, Taufkirchen, Germany) for 5 min at 37 °C. Subsequently, the hMSCs were collected and washed twice with RPMI/10% FCS. Subconfluently growing hMSCs were incubated at 37 °C in the presence or absence of different concentrations of silver nanoparticles for 24 h under cell culture conditions.The viability and the morphology of the incubated hMSCs were analysed using calcein-acetoxymethylester (calcein-AM, Calbiochem, Schwalbach, Germany) fluorescence staining. After incubation for 24 h, the nanoparticle-treated cells and silver ion-treated cells were washed twice with RPMI and incubated with calcein-AM (1 μM) at 37 °C for 30 min under cell culture conditions. Subsequently, the adherent cells were washed again with RPMI and analysed by fluorescence microscopy (Olympus MVX10, Olympus, Hamburg, Germany). Fluorescence microphotographs were taken (Cell P, Olympus) and digitally processed using Adobe Photoshop® 7.0. The cell morphology was analysed by phase-contrast microscopy (Olympus MVX). In this method, living cells give rise to green fluorescence.
Polymorphonuclear neutrophil granulocytes (PMNs), which are known to be potent producers of ROS, were isolated by using a single-step procedure that was based on a discontinuous double-Ficoll gradient described by English and Andersen.26 Cell counting was performed using Tuerk staining solution (Sigma-Aldrich). The isolated cells were adjusted to 1 × 106 cells mL−1 in RPMI cell culture medium supplemented with L-glutamine (0.3 g L−1), sodium bicarbonate (2.0 g L−1), 10% (v/v) FCS, and 20 mM N-(2-hydroxyethyl)-piperazine-N′-(2-ethanesulfonic acid) (HEPES, Sigma-Aldrich).
To monitor the generation of ROS from silver-treated PMNs, the oxidant-sensitive dye, DCFH-DA (2′,7′-dichlorofluorescein-diacetate), was used, as described by Hsin et al.27 and Carlson et al.28 After 1 h exposure to Ag-NPs or silver ions or 10 μM fMLP (formyl-methionyl-leucyl-phenylalanine), 10 min before DCFH-DA addition, prior to analysis, as a positive control, the cells were centrifuged at 370g for 5 min at room temperature. The supernatants were discarded, and the pellets were carefully resuspended. Subsequently, the cells were incubated with 20 μM DCFH-DA (resuspended in RPMI/10% FCS) for 30 min in the dark (37 °C, 5% CO2). After incubation, the cells were washed, centrifuged and fixed with 1.5% paraformaldehyde. The fluorescent signal was monitored by flow cytometry.
Bacterial culture experiments
Bacterial tests were performed with Staphylococcus aureus (DSMZ 1104) obtained from the DSMZ (German Collection of Microorganisms and Cell Cultures). S. aureus was grown in BHI broth (brain-heart infusion broth, bioMérieux, Nürtingen, Germany) overnight at 37 °C in a water bath. Bacterial concentrations of overnight cultures were measured using a Densichek® (bioMérieux, Lyon, France) turbidity photometer. The calculation of bacterial counts was based on turbidity standard solutions (McFarland scale).The antimicrobial activity of nanoparticulate silver was tested using standard methods which determine the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC). The MIC was determined in RPMI 1640 (Life Technologies) containing 10% (v/v) fetal calf serum (FCS, Life Technologies) and L-glutamine (0.3 g L−1, Life Technologies), and defined as the lowest silver concentration required to inhibit bacterial growth (no visible growth) in 2 mL plastic macrodilution test tubes. Therefore, working silver stock solutions (50 μL) were added to 1 mL of the respective liquid culture medium, and different cell numbers (103 to 106 mL−1) of bacteria were used for inoculation. Cells were incubated in a cell culture incubator (RPMI/10% FCS) in the presence of 5% carbon dioxide in a humidified atmosphere at 37 °C overnight.
The minimum bactericidal concentration (MBC) was subsequently determined by plating 100 μL aliquots of the MIC samples on blood agar plates. The MBC was defined as the lowest silver concentration that completely prevented colony forming units (CFU) on the agar plate.
Statistical analysis
Data are expressed as mean ± standard deviation of at least three independent experiments. Analysis of the data distribution was performed using Student's t-test to analyse the significance of differences between the treated group and the control group (without silver exposure). P values of less than 0.05 were considered statistically significant.Results and discussion
Dissolution experiments of silver nanoparticles
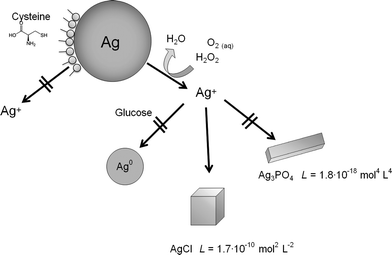
The dissolution of silver nanoparticles was monitored by long-term experiments using dialysis bags containing nanoparticles which were permeable only for silver ions. Previous studies have shown that the dissolution is fast at the beginning and slows down over time, leading to incompletely dissolved particles.15 Of course, an immersion in pure water is far from realistic conditions in biological or environmental media. As these media are very complex in nature and many components will act simultaneously on the silver nanoparticles, we have simplified the system by the addition of various model compounds. We have added various compounds that are also present in biological media to the immersion medium and measured the dissolution curves. In particular, we added chloride and phosphate as typical anions that can lead to the precipitation of sparingly soluble silver salts (AgCl and Ag3PO4), glucose as a reducing sugar, and cysteine as a model for sulphur-containing proteins. As dissolved oxygen is the only agent that can dissolve metallic silver under these conditions, we also varied the oxygen content of the solution. Finally, H2O2 was used as a strongly oxidizing compound.Representative dissolution curves for PVP-coated silver nanoparticles are shown in Fig. 1.
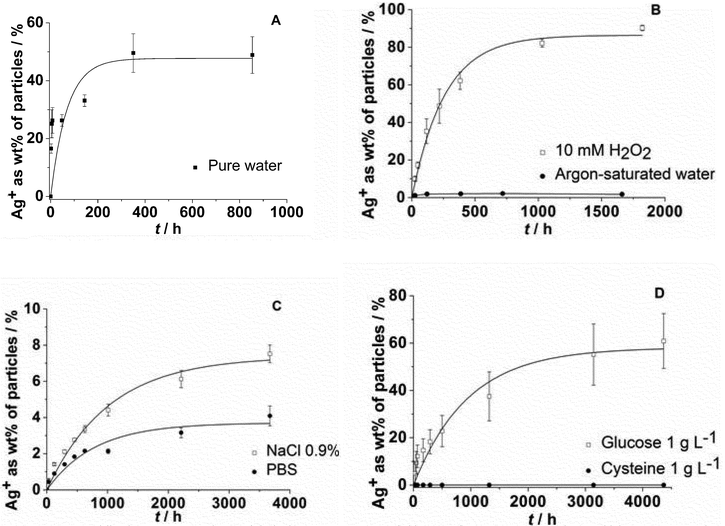 | ||
| Fig. 1 Dissolution of silver nanoparticles immersed in pure water (A: data taken from ref. 15), degassed, argon-saturated water under an argon atmosphere, and in water with 10 mM H2O2 (B), in aqueous NaCl (0.9%) or in PBS (C), and in the presence of either cysteine (aq.: 1 g L−1) or glucose (aq.: 1 g L−1) (D). | ||
The dissolution requires the presence of dissolved oxygen as it is obvious from Fig. 1B. If no oxygen is present, only a very small fraction of silver is dissolved, possibly by traces of oxygen in the experimental setup. An oxidizing agent like H2O2 clearly enhances the dissolution. The presence of dissolved NaCl, either in pure form or as PBS, strongly slows down the dissolution (Fig. 1C), probably due to the formation of silver chloride on the particle surface (passivation). Cysteine has a clearly inhibiting effect with almost no dissolution of the silver nanoparticles. Glucose has a decelerating effect but leads to a similar final dissolved fraction (Fig. 1D). This suggests that cysteine adsorbs onto the silver nanoparticle surface with its thiol group and prevents the subsequent oxidation of silver. In contrast, glucose slows down the dissolution, but clearly does not prevent the oxidation on a longer time scale.
Incomplete dissolution of nanoparticles
The fact that silver nanoparticles dissolve only partially in the presence of air unless a very strong oxidizing agent like H2O2 is present is astonishing, but corresponds to previous results by us15 and Zhang et al.21 We will discuss this observation in the following paragraph.There appears to be agreement in the literature that dissolved oxygen is the oxidizing species for silver under normal circumstances. A complexation by organic ligands may facilitate the oxidation of silver by shifting the equilibrium, but cannot explain the dissolution by itself. Another remarkable observation is the only partial dissolution of silver nanoparticles, even after a long time.15 This may be due to a lack of the oxidizing species, i.e. oxygen. We have therefore measured the oxygen concentration in the air-saturated surrounding water and found to be 5.2 mg O2 per L, i.e. 160 μM O2. This is more than enough to oxidize 1.4 mg of metallic silver in the system (13 μmol dissolved in 400 mL, i.e. 32 μM). We have also performed the dissolution in pure water for 500 h and then exchanged the surrounding medium by fresh water. The dissolution starts again, but not as much as expected and leads to a new “equilibrium” (Fig. 2).
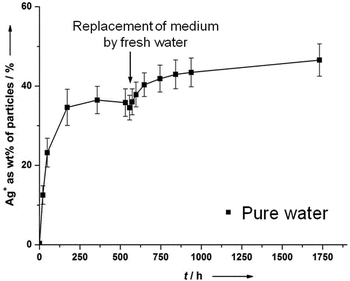 | ||
| Fig. 2 Partial dissolution of silver nanoparticles (PVP-coated; 70 nm), immersed in pure water, with exchange of the surrounding medium by fresh water. The access of oxygen was allowed at all times. | ||
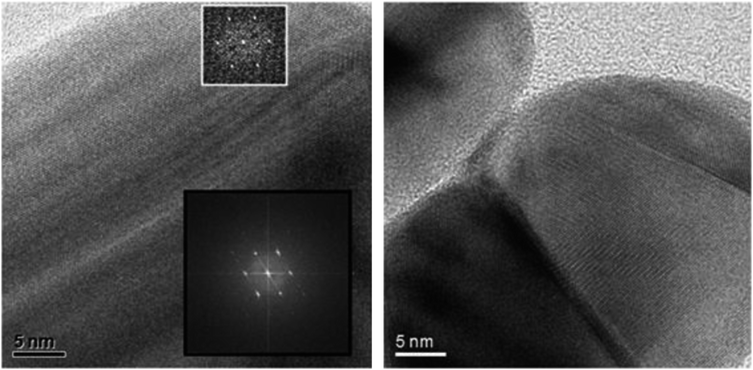
This demonstrates that it is not the presence of oxygen alone which leads to a dissolution of silver. An explanation might be a kind of surface passivation during immersion. High-resolution transmission electron microscopy on immersed silver nanoparticles was therefore performed to investigate the presence of any passivating surface layer after immersion in water. Fig. 3 shows representative images. A change of the nanoparticle surface was never detectable, ruling out the presence of a surface passivation.
Behaviour of silver nanoparticles in complex media
In order to better understand the processes during immersion, we have extended these studies by mixing silver nanoparticle dispersions with media of increasingly biological nature and studying the (nano-)particulate reaction products. The solutions/dispersions were stirred for equilibration and then subjected to ultracentrifugation. All precipitates and nanoparticles were isolated by this way and then analysed by scanning electron microscopy, energy-dispersive X-ray spectroscopy and X-ray powder diffraction. This gives an integral view on all the particulate species that are present after long-term immersion. The results are summarized in Table 2. The original silver nanoparticles were found practically unchanged after the immersion, indicating that it had only partially dissolved.| Medium | Particles found in the ultracentrifugate, with diameter | Maximum possible concentrations of Ag+ in the immersion medium/μg L−1 | Concentration of Cl−/M |
|---|---|---|---|
| Glucose (2 g L−1) | Ag (70 nm) | 0 | |
| NaCl (0.9%) | Ag (70 nm), AgCl (400–800 nm) | 0.225 | 0.154 |
| NaCl (0.9%) + glucose (2 g L−1) | Ag (70 nm), AgCl (200–500 nm) | 0.225 | 0.154 |
| PBS | Ag (70 nm), AgCl (200–300 nm) | 0.250 | 0.139 |
| PBS + glucose (2 g L−1) | Ag (70 nm), AgCl (200–500 nm) | 0.250 | 0.139 |
| RPMI 1640 medium/10% FCS | Ag (70 nm), AgCl (110–140 nm) | 0.327 | 0.108 |
| LB medium | Ag (70 nm), AgCl (250–300 nm) | 0.372 | 0.085 |
The results show that the released silver ions are mainly precipitated as AgCl if chloride is present. We have also computed the maximum concentration of silver in the presence of various inorganic ions and found that less than one μg of silver per litre can be present in thermodynamic equilibrium. This, of course, neglects the influence of organic species that may act as coordinating ligands and also a possible non-equilibrium state of the dispersion. The initially present silver nanoparticles were recovered in all cases. Silver phosphate was never observed, probably due to the moderate pH (around 7) at which phosphate is mostly protonated to hydrogen phosphate and dihydrogen phosphate.
Cell and bacterial culture experiments with silver nanoparticles stored under air and under argon
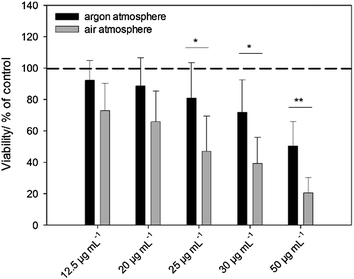
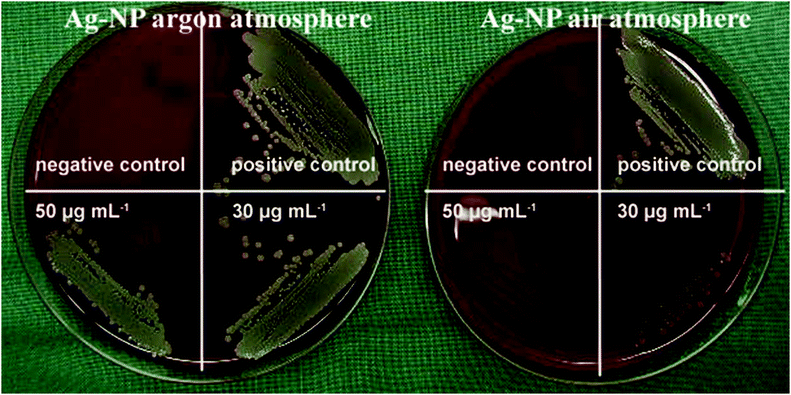
Our hypothesis that the release of silver ions determines silver toxicity was confirmed by cell culture experiments. If the nanoparticles were stored under argon, they did not partially dissolve and therefore were much less toxic towards tissue-like cells than the same particles stored under air (Fig. 4). In addition, the same results were obtained by microbiological experiments using S. aureus where oxidized silver nanoparticles were much more toxic than those stored under air (Fig. 5 and Table 3). Of course, the particles will dissolve after coming into contact with air, therefore they will become increasingly bactericidal (and cytotoxic) with time.| Number of bacteria used for inoculation | Ag nanoparticles stored under an argon atmosphere | Ag nanoparticles stored under air |
|---|---|---|
| 1 × 106 | >50 | >50 |
| 1 × 105 | >50 | 50 |
| 1 × 104 | >50 | 30–25 |
| 1 × 103 | 25 | 25–20 |
Formation of reactive oxygen species (ROS)
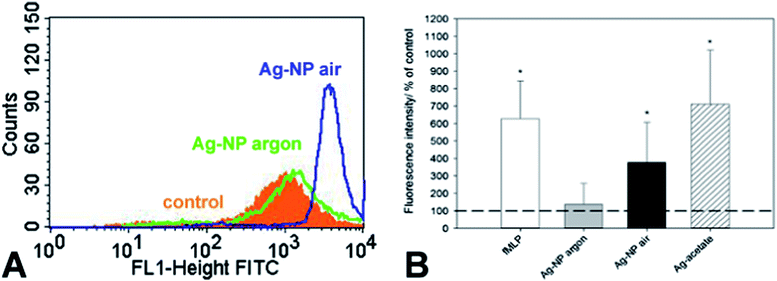
We and others have shown that reactive oxygen species (ROS) were generated from cells as a consequence of silver nanoparticle exposure and that the formation of ROS contributes to silver induced cell toxicity.29–32 Air-stored silver nanoparticles induced a pronounced increase in cellular ROS formation compared to the same amount of argon-stored silver nanoparticles (Fig. 6A and B). The addition of silver acetate (the same silver concentration as the nanoparticles) induced the highest release in ROS under these conditions comparable to the standard cell stimulus formyl-methionyl-leucyl-phenylalanine (fMLP).Silver nanoparticles enter the cell by endocytosis and end up first in an endosome and then in a lysosome.33 There, they encounter acidic conditions with a pH between 4 and 5.34 Liu and Hurt have studied the dissolution of silver nanoparticles at different pH and found an increase of the ion release at lower pH. We can therefore assume that the silver nanoparticles will undergo accelerated dissolution after endosomal uptake. However, the time in the cell culture medium is much longer than the time in the lysosome, therefore the toxic action of silver is probably due to both effects.
A model for the behaviour of silver nanoparticles in complex media
The typically applied methods for silver determination are AAS or ICP-MS, and the separation of silver nanoparticles and silver ions is typically accomplished by dialysis or nanofiltration. In general, although there are a number of studies where the dissolution kinetics of dispersed silver nanoparticles was measured, they are practically incomparable (Table 1). The properties of the nanoparticles were rather diverse (e.g. size, charge, functionalization), and it is very likely that this strongly influences their dissolution behaviour. Furthermore, the immersion media are different and not easily comparable between the individual data points. Finally, it can be safely assumed that the total silver concentration will influence the dissolution process, as critical factors like the solubility product of silver chloride or the reduction potential of silver ions directly depend on the concentration. However, some conclusions can be drawn, mainly by comparing results within one set of experiments, i.e. from one reference.Liu et al. described the dissolution of silver nanoparticles in acetate buffer in the presence of citrate anions, sodium sulphide and 11-mercaptoundecanoic (MUA) acid and found that sulphidation and MUA reduced the dissolution rate to undetectable values.13 They also investigated the role of humic and fulvic acids, dissolved oxygen, natural and low salt sea water and showed that the addition of organic matter can suppress the release of silver ions.12 Levard et al. investigated the role of a surface sulphidation process and found that at a high ratio of sulphur to silver, the released silver content decreased to hardly detectable values.18 Zook et al. measured the dissolution in cell culture medium (DMEM) and found a fractional dissolution of different polymer coated silver nanoparticles. The dissolution was enhanced in biological media in comparison to inorganic salt solutions which is probably due to complexation of the released silver ions.20 Zhang et al. considered size effects on the dissolution kinetics in quarter-strength Hoagland medium and concluded the dependence of size and concentration on the release rate. Smaller particles were found to dissolve faster.21 The fate of silver nanoparticles in synthetic gastric fluid and wound fluid was studied by Liu et al. They reported a rapid dissolution in gastric acid in comparison to pseudoextracellular fluid and also found an interaction with thiol- and selenide-containing biomolecules. They proposed the formation of secondary thiol- and selenide-containing nanoparticles after partial dissolution in the gastrointestinal tract.23
Our own observations and the collected literature data led us to the model that is depicted in Fig. 7.
Conclusions
From the reported experiments and the available literature on the dissolution of silver nanoparticles, we conclude that(1) Metallic silver is oxidized by dissolved molecular oxygen.
(2) Thiol- or selenide-containing compounds can block the particle surface and prevent the dissolution (in our studies represented by a cysteine molecule).
(3) Once silver ions are released into the solution, they are rapidly precipitated as silver chlorides, apparently not on the surface of the initial silver particles, but as separate particles. The equilibrium concentration of Ag+ is very small (<1 ppb).
(4) Silver phosphate is not formed if chloride is present.
(5) Reducing sugars (here: glucose) slow down, but do not prevent the oxidative dissolution of silver.
(6) Organic molecules can lead to a complexation of silver ions and thereby accelerate the dissolution.
(7) Silver nanoparticles do not dissolve completely in the presence of molecular oxygen. A stronger oxidation agent like H2O2 is necessary for a complete dissolution.
(8) The toxic species for eukaryotic cells and for bacteria are silver ions that are released after oxidation of silver nanoparticles. These lead to the formation of reactive oxygen species (ROS).
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft (DFG) for financial support of this project within the Priority Program NanoBio Responses (SPP1313). We thank the network of excellence agening.net (CS2010-11384-E) for generous support.Notes and references
- Y. Teow, P. V. Asharani, M. P. Hande and S. Valiyaveettil, Chem. Commun., 2011, 47, 7025–7038 RSC.
- B. Nowack, H. F. Krug and M. Height, Environ. Sci. Technol., 2011, 45, 1177–1183 CrossRef CAS PubMed.
- F. Gottschalk and B. Nowack, J. Environ. Monit., 2011, 13, 1145–1155 RSC.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Molecules, 2011, 16, 8894–8918 CrossRef CAS PubMed.
- B. Simoncic and B. Tomsic, Text. Res. J., 2010, 80, 1721–1737 CrossRef CAS PubMed.
- K. Chaloupka, Y. Malam and A. M. Seifalian, Trends Biotechnol., 2010, 28, 580–588 CrossRef CAS PubMed.
- J. W. Alexander, Surg. Infect.: Sel. Antibiot. Ther., 2009, 10, 289–292 CrossRef PubMed.
- X. Chen and H. J. Schluesener, Toxicol. Lett., 2008, 176, 1–12 CrossRef CAS PubMed.
- G. A. Sotiriou and S. E. Pratsinis, Curr. Opin. Chem. Eng., 2011, 1, 3–10 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- Z. M. Xiu, Q. B. Zhang, H. L. Puppala, V. L. Colvin and P. J. J. Alvarez, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- L. Liu and R. H. Hurt, Environ. Sci. Technol., 2010, 44, 2169–2175 CrossRef PubMed.
- J. Liu, D. A. Sonshine, S. Shervani and R. H. Hurt, ACS Nano, 2010, 4, 6903–6913 CrossRef CAS PubMed.
- X. Li, J. J. Lenhart and H. W. Walker, Langmuir, 2010, 26, 16690–16698 CrossRef CAS PubMed.
- S. Kittler, C. Greulich, J. Diendorf, M. Köller and M. Epple, Chem. Mater., 2010, 22, 4548–4554 CrossRef CAS.
- C. M. Ho, S. K. W. Yau, C. N. Lok, M. H. So and C. M. Che, Chem. – Asian J., 2010, 5, 285–293 CrossRef CAS PubMed.
- G. A. Sotiriou and S. E. Pratsinis, Environ. Sci. Technol., 2010, 44, 5649–5654 CrossRef CAS PubMed.
- C. Levard, B. C. Reinsch, F. M. Michel, C. Oumahi, G. V. Lowry and G. E. Brown, Environ. Sci. Technol., 2011, 45, 5260–5266 CrossRef CAS PubMed.
- C. M. Ho, C. K. Wong, S. K. W. Yau, C. N. Lok and C. M. Che, Chem. – Asian J., 2011, 6, 2506–2511 CrossRef CAS PubMed.
- J. M. Zook, S. E. Long, D. Cleveland, C. L. A. Geronimo and R. I. MacCuspie, Anal. Bioanal. Chem., 2011, 401, 1993–2002 CrossRef CAS PubMed.
- W. Zhang, Y. Yao, N. Sullivan and Y. Chen, Environ. Sci. Technol., 2011, 45, 4422–4428 CrossRef CAS PubMed.
- Z. M. Xiu, J. Ma and P. J. J. Alvarez, Environ. Sci. Technol., 2011, 45, 9003–9008 CrossRef CAS PubMed.
- J. Y. Liu, Z. Y. Wang, F. D. Liu, A. B. Kane and R. H. Hurt, ACS Nano, 2012, 6, 9887–9899 CrossRef CAS PubMed.
- H. Wang, X. Qiao, J. Chen and S. Ding, Colloids Surf., A, 2005, 256, 111–115 CrossRef CAS PubMed.
- C. N. Lok, C. M. Ho, R. Chen, Q. Y. He, W. Y. Yu, H. Sun, P. K. H. Tam, J. F. Chiu and C. M. Che, J. Biol. Inorg. Chem., 2007, 12, 527–534 CrossRef CAS PubMed.
- D. English and B. R. Andersen, J. Immunol. Methods, 1974, 5, 249–252 CrossRef CAS.
- Y. H. Hsin, C. F. Chen, S. Huang, T. S. Shih, P. S. Lai and P. J. Chueh, Toxicol. Lett., 2008, 179, 130–139 CrossRef CAS PubMed.
- C. Carlson, S. M. Hussain, A. M. Schrand, L. K. Braydich-Stolle, K. L. Hess, R. L. Jones and J. J. Schlager, J. Phys. Chem. B, 2008, 112, 13608–13619 CrossRef CAS PubMed.
- O. Choi and Z. Q. Hu, Environ. Sci. Technol., 2008, 42, 4583–4588 CrossRef CAS.
- H. J. Park, J. Y. Kim, J. Kim, J. H. Lee, J. S. Hahn, M. B. Gu and J. Yoon, Water Res., 2009, 43, 1027–1032 CrossRef CAS PubMed.
- C. Greulich, J. Diendorf, J. Geßmann, T. Simon, T. Habijan, G. Eggeler, T. A. Schildhauer, M. Epple and M. Köller, Acta Biomater., 2011, 7, 3505–3514 CrossRef CAS PubMed.
- D. Guo, L. Zhu, Z. Huang, H. Zhou, Y. Ge, W. Ma, J. Wu, X. Zhang, X. Zhou, Y. Zhang, Y. Zhao and N. Gu, Biomaterials, 2013, 34, 7884–7894 CrossRef CAS PubMed.
- C. Greulich, J. Diendorf, T. Simon, G. Eggeler, M. Epple and M. Köller, Acta Biomater., 2011, 7, 347–354 CrossRef CAS PubMed.
- P. R. Gil, M. Nazarenus, S. Ashraf and W. J. Parak, Small, 2012, 8, 943–948 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2014 |