Integration of a CRISPR Cas12a-assisted multicolor biosensor and a micropipette tip enables visible point-of-care testing of foodborne Vibrio vulnificus†
Abstract
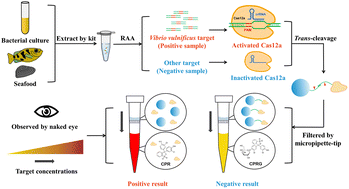
Foodborne pathogens cause numerous food safety problems, and as a virulent bacterium falling under this category, Vibrio vulnificus (V. vulnificus) poses a huge threat to public health. The conventional methods used for the detection of V. vulnificus, including culture-based and molecular detection methods, have a variety of drawbacks, including being time-consuming and labor-intensive, the requirement of large-scale equipment, and the lack of professional operators. This paper establishes a visible detection platform for V. vulnificus based on CRISPR/Cas12a, which is integrated with nucleic acid isothermal amplification and β-galactosidase-catalyzed visible color reaction. The specific vvhA gene and a conservative segment in the 16S rDNA gene of the Vibrio genus were selected as the detection targets. By using spectrum analysis, this CRISPR detection platform achieved sensitive detection of V. vulnificus (1 CFU per reaction) with high specificity. Through the color transformation system, as low as 1 CFU per reaction of V. vulnificus in both bacterial solution and artificially contaminated seafood could be visibly observed with the naked eye. Furthermore, the consistency between our assay and the qPCR assay in the detection of V. vulnificus spiked seafood was confirmed. In general, this visible detection platform is user-friendly, accurate, portable, and equipment-free, and is expected to provide a powerful supplement in point-of-care testing of V. vulnificus and also holds good promise for future application in foodborne pathogen detection.
- This article is part of the themed collection: Analyst HOT Articles 2023


 Please wait while we load your content...
Please wait while we load your content...