Linear dichroism as a probe of molecular structure and interactions
Abstract
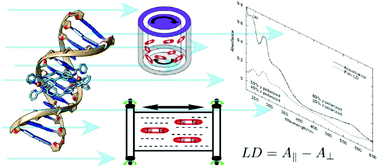
Linear dichroism (LD) spectroscopy involves measuring the wavelength (or energy) dependence of the difference in absorption of light parallel and perpendicular to an orientation direction. It requires samples to have a net orientation. The aim of this review is to summarise some UV–visible linear dichroism (LD) methods that can be usefully applied to increase our understanding of biomacromolecules and their complexes that have a high aspect ratio. LD shares the advantages of most spectroscopic techniques including the fact that data collection is fairly straightforward and many sample types can be investigated. Conversely, LD shares the disadvantage that the measured signal is an average over all species in the sample on which the light beam is incident. LD mitigates this disadvantage somewhat in that only species which are oriented give a net signal. How the data can be analysed to give structural information about small molecules in stretched films and membrane systems or bound to biomacromolecules and directly about biomacromolecules such as DNA and protein fibres forms part of this review. In the UV–visible region LD often suffers noticeably from light scattering since the samples tend to be large relative to the wavelength of the incident light, so consideration is also given to data analysis challenges including removal of scattering contributions to an observed signal. Brief mention is made of fluorescence detected LD.


 Please wait while we load your content...
Please wait while we load your content...