Insights into the binding modes of CC chemokine receptor 4 (CCR4) inhibitors: a combined approach involving homology modelling, docking, and molecular dynamics simulation studies†
Abstract
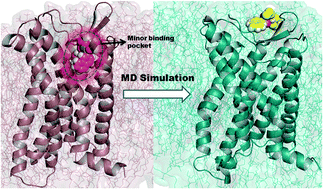
CC chemokine receptor 4 (CCR4), a G protein-coupled receptor (GPCR), plays a vital role in the progression of asthma, T-cell lymphoma, inflammation, and Alzheimer's disease. To date, the structure of CCR4 has not been determined. Therefore, the nature of the interactions between inhibitors and CCR4 is not well known. In this study, we used CCR5 as a template to model the structure of CCR4. Docking studies were performed for four naphthalene-sulphonamide derivatives and crucial ligand–protein interactions were analysed. Molecular dynamics (MD) simulations of these complexes (100 ns each) were carried out to gain insights into the interactions between ligands and CCR4. MD simulations revealed that the residues identified by the docking were displaced and new residues were inserted near the ligands. Results of a principal component analysis (PCA) suggested that CCR4 unfolds at the extracellular site surrounding the ligands. Our simulations identified crucial residues involved in CCR4 antagonism, which were supported by previous mutational studies. Additionally, we identified Ser3.29, Leu3.33, Ser5.39, Phe6.47, Ile7.35, Thr7.38, Thr7.40, and Ala7.42 as residues that play crucial roles in CCR4 antagonism. Mutational studies will help elucidate the significance of these residues in CCR4 antagonism. An understanding of ligand–CCR4 interactions might aid in the design of novel CCR4 inhibitors.

 Please wait while we load your content...
Please wait while we load your content...