An entropy-like index of bifurcational robustness for metabolic systems†
Abstract
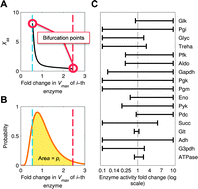
Natural and synthetic metabolic pathways need to retain stability when faced against random changes in gene expression levels and kinetic parameters. In the presence of large parameter changes, a robust system should specifically avoid moving to an unstable region, an event that would dramatically change system behavior. Here we present an entropy-like index, denoted as S, for quantifying the bifurcational robustness of metabolic systems against loss of stability. We show that S enables the optimization of a metabolic model with respect to both bifurcational robustness and experimental data. We then demonstrate how the coupling of ensemble modeling and S enables us to discriminate alternative designs of a synthetic pathway according to bifurcational robustness. Finally, we show that S enables the identification of a key enzyme contributing to the bifurcational robustness of yeast glycolysis. The different applications of S demonstrated illustrate the versatile role it can play in constructing better metabolic models and designing functional non-native pathways.
- This article is part of the themed collection: Integrative Approaches for Signalling and Metabolic Networks

 Please wait while we load your content...
Please wait while we load your content...