The pan-cancer analysis of gene expression patterns in the context of inflammation†
Abstract
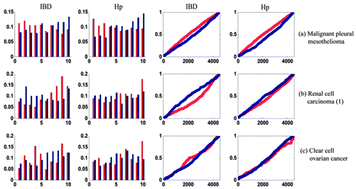
Although several studies have investigated the essential roles of inflammation in tumor progression, not many have systematically analyzed gene expression patterns across diverse cancers in the context of inflammation. In this study, in order to better understand the inflammatory scenario, we initially constructed the inflammatory timeline (IT) based on two gene expression profiles during inflammatory progression (inflammatory bowel disease and Helicobacter pylori infection). Then, we separately identified the differentially expressed genes (DEGs) from 25 cancer-related microarray data. By comparing the distributions of DEGs in the IT, we identified three novel pan-cancer gene expression patterns. In the first pattern, the up-regulated genes in cancers were over-expressed in the early phase of inflammation, while the down-regulated genes were over-expressed in the late phase of inflammation. The second pattern was the opposite of the first one. The third pattern appeared to be transitional between the first and second patterns. We found that some cancers with different tissue origins have similar gene expression patterns. Finally, we identified two sets of tissue-independent inflammatory signatures that were over-expressed in early and late phases of inflammation, respectively. The dominant biological processes of early inflammatory signatures were cell proliferation, DNA replication, and DNA repair, whereas the late inflammatory signatures were reflective of innate immune response, neutrophil migration, and antigen processing. These inflammatory signatures may be useful to predict gene expression patterns in human cancers. Therefore, the pan-cancer analysis of gene expression patterns in the context of inflammation provides a novel insight into cancers and an unprecedented opportunity to develop new therapies.

 Please wait while we load your content...
Please wait while we load your content...