Sizing up single-molecule enzymatic conformational dynamics
Abstract
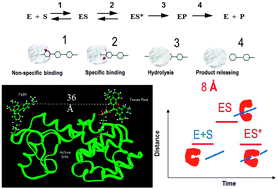
Enzymatic reactions and related protein conformational dynamics are complex and inhomogeneous, playing crucial roles in biological functions. The relationship between protein conformational dynamics and enzymatic reactions has been a fundamental focus in modern enzymology. It is extremely difficult to characterize and analyze such complex dynamics in an ensemble-averaged measurement, especially when the enzymes are associated with multiple-step, multiple-conformation complex chemical interactions and transformations. Beyond the conventional ensemble-averaged studies, real-time single-molecule approaches have been demonstrated to be powerful in dissecting the complex enzymatic reaction dynamics and related conformational dynamics. Single-molecule enzymology has come a long way since the early demonstrations of the single-molecule spectroscopy studies of enzymatic dynamics about two decades ago. The rapid development of this fundamental protein dynamics field is hand-in-hand with the new development of single-molecule imaging and spectroscopic technology and methodology, theoretical model analyses, and correlations with biological preparation and characterization of the enzyme protein systems. The complex enzymatic reactions can now be studied one molecule at a time under physiological conditions. Most exciting developments include active manipulation of enzymatic conformational changes and energy landscape to regulate and manipulate the enzymatic reactivity and associated conformational dynamics, and the new advancements have established a new stage for studying complex protein dynamics beyond by simply observing but by actively manipulating and observing the enzymatic dynamics at the single-molecule sensitivity temporally and spatially.
- This article is part of the themed collection: Single-molecule optical spectroscopy

 Please wait while we load your content...
Please wait while we load your content...