A G-quadruplex DNAzyme chemiluminescence aptasensor based on the target triggered DNA recycling for sensitive detection of adenosine†
Abstract
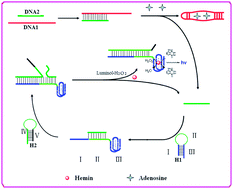
A G-quadruplex DNAzyme chemiluminescence aptasensor based on the target triggered DNA recycling was constructed for sensitive detection of adenosine. The system involves four nucleic acid strands, namely DNA1, DNA2, H1 and H2. DNA1 is a chimeric conjugate of the adenosine aptamer strand, DNA2 is an inhibitory strand that contains complementary sequences to DNA1. Hairpin structures H1 and H2 are designed according to the sequences of DNA2. In the absence of adenosine, DNA1 is inhibited by hybridizing with DNA2, and H1 and H2 coexist in solution. In the presence of adenosine, the DNA1 strand binds adenosine, and results in the release of DNA2. Upon interaction of DNA2 with H1, the hairpin structure of H1 is opened, resulting in the activation of the G-quadruplex. Meanwhile, the unfolded region in H1 can serve as a toehold to hybridize with H2, leading to the displacement of the DNA2 through a branch migration process. Thus, the DNA2 dissociates from the H1–H2 complex. The released DNA2 hybridizes with another H1 and initiates the second cycle, and generates a number of H1–H2 complexes containing the G-riched DNA. The G-riched DNA assembles with hemin to form the hemin–G-quadruplexes that exhibit peroxidase-like activity which can catalyze oxidation of luminol by H2O2 generating chemiluminescence. In this way, a single adenosine can trigger many H1–H2 complexes and activate many G-quadruplex DNAzymes, which leads to the significant chemiluminescence enhancement. The linear detection range of adenosine with the proposed aptasensor is 0.8 to 200 μM, and the detection limit comes down to 0.6 μM. Most importantly, no organic dye molecule-labeled DNA or extra nanomaterials were used in this proposed CL aptasensor. The method is simple, sensitive and inexpensive and has potential for application in clinical diagnostic assays.

 Please wait while we load your content...
Please wait while we load your content...