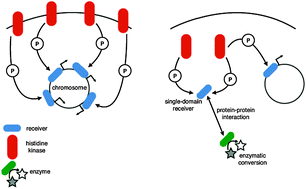
A major mode of signal transduction in bacteria is the two-component system, which involves phosphorylation of an output-generating receiver protein by a signal-sensing histidine kinase. This differs from the more common one-component system—where both signal sensing and output generation are performed by the same protein—in the spatial separation of the two activities and the obligate need for post-translational modification (phosphorylation). Many described two-component systems involve a linear structure where a single kinase phosphorylates a cognate receiver. However, inherently branched network structures are being increasingly discovered, though their prevalence is unknown. Though the simpler one-component systems are more common than two-component systems, some organisms encode a disproportionately high number of the latter; though these organisms are generally described as having ‘complex’ lifestyles, no systematic description of their signaling networks has been proposed. Finally, the relative contributions of the two modes of signal transduction towards achieving an optimal regulatory cost for growth and survival in an environment remain poorly understood. Here we present a comparative genomics survey of ∼165 000 regulatory proteins from ∼850 prokaryotic genomes and suggest that organisms with elevated occurrence of two-component systems—which generally belong to phylogenetic classes with relatively poor representation in genomic databases—also code for more complex and branched two-component networks. Such branched signaling might compensate for the apparent paucity in the total number of regulatory proteins these organisms encode. Finally, such interconnected signaling networks might be more common than anticipated, indicating the pressing need for genome-scale experimental studies of signaling networks in many understudied phylogenetic groups of organisms.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...