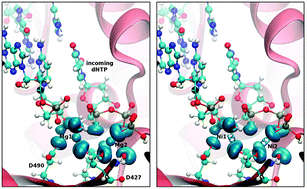
DNA polymerases require two divalent metal ions in the active site for catalysis. Mg2+ has been confirmed to be the most probable cation utilized by most polymerases in vivo. Other metal ions are either potent mutagens or inhibitors. We used structural and topological analyses based on ab initioQM/MM calculations to study human DNA polymerase λ (Polλ) with different metals in the active site. Our results indicate a slightly longer O3′–Pα distance (∼3.6 Å) for most inhibitor cations compared to the natural and mutagenic metals (∼3.3–3.4 Å). Optimization with a larger basis set for the previously reported transition state (TS) structures (Cisneros et al., DNA Repair, 2008, 7, 1824.) gives barriers of 17.4 kcal mol−1 and 15.1 kcal mol−1 for the Mg2+ and Mn2+ catalyzed reactions respectively. Relying on the key relation between the topological signature of a metal cation and its selectivity within biological systems (de Courcy et al., J. Chem. Theor. Comput., 2010, 6, 1048.) we have performed electron localization function (ELF) topological analyses. These analyses show that all inhibitor and mutagenic metals considered, except Na+, present a “split” of the outer-shell density of the metal. This “splitting” is not observed for the non-mutagenic Mg2+ metal. Population and multipole analyses on the ELF basins reveal that the electronic dipolar and quadrupolar polarization is significantly different with Mg2+ compared to all other cations. Our results shed light at the atomic level on the subtle differences between Mg2+, mutagenic, and inhibitor metals in DNA polymerases. These results provide a correlation between the electronic distribution of the cations in the active site and the possible consequences on DNA synthesis.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...