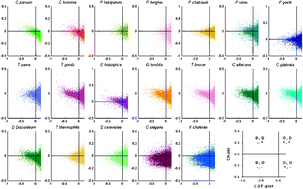
Parasitic protozoal infections have long been known to cause profound degrees of sickness and death in humans as well as animal populations. Despite the increase in the number of annotated genomes available for a large variety of protozoa, a great deal more has yet to be learned about them, from their fundamental physiology to mechanisms invoked during host–pathogen interactions. Most of these genomes share a common feature, namely a high prevalence of low complexity regions in their predicted proteins, which is believed to contribute to the uniqueness of the individual species within this diverse group of early-branching eukaryotes. In the case of Plasmodium species, which cause malaria, such regions have also been reported to hamper the identification of homologues, thus making functional genomics exceptionally challenging. One of the better accepted theories accounting for the high number of low complexity regions is the presence of intrinsic disorder in these microbes. In this study we compare the degree of disordered proteins that are predicted to be expressed in many such ancient eukaryotic cells. Our findings indicate an unusual bias in the amino acids comprising protozoal proteomes, and show that intrinsic disorder is remarkably abundant among their predicted proteins. Additionally, the intrinsically disordered regions tend to be considerably longer in the early-branching eukaryotes. An analysis of a Plasmodium falciparum interactome indicates that protein–protein interactions may be at least one function of the intrinsic disorder. This study provides a bioinfomatics basis for the discovery and analysis of unfoldomes (the complement of intrinsically disordered proteins in a given proteome) of early-branching eukaryotes. It also provides new insights into the evolution of intrinsic disorder in the context of adapting to a parasitic lifestyle and lays the foundation for further work on the subject.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...