A cell–ECM screening method to predict breast cancer metastasis†
Abstract
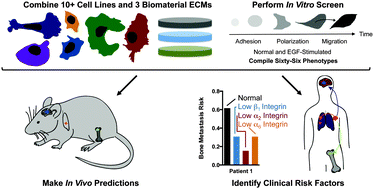
Breast cancer preferentially spreads to the bone, brain, liver, and lung. The clinical patterns of this tissue-specific spread (tropism) cannot be explained by blood flow alone, yet our understanding of what mediates tropism to these physically and chemically diverse tissues is limited. While the microenvironment has been recognized as a critical factor in governing metastatic colonization, the role of the extracellular matrix (ECM) in mediating tropism has not been thoroughly explored. We created a simple biomaterial platform with systematic control over the ECM protein density and composition to determine if integrin binding governs how metastatic cells differentiate between secondary tissue sites. Instead of examining individual behaviors, we compiled large patterns of phenotypes associated with adhesion to and migration on these controlled ECMs. In combining this novel analysis with a simple biomaterial platform, we created an in vitro fingerprint that is predictive of in vivo metastasis. This rapid biomaterial screen also provided information on how β1, α2, and α6 integrins might mediate metastasis in patients, providing insights beyond a purely genetic analysis. We propose that this approach of screening many cell–ECM interactions, across many different heterogeneous cell lines, is predictive of in vivo behavior, and is much simpler, faster, and more economical than complex 3D environments or mouse models. We also propose that when specifically applied toward the question of tissue tropism in breast cancer, it can be used to provide insight into certain integrin subunits as therapeutic targets.

 Please wait while we load your content...
Please wait while we load your content...