Quantitative proteomics and systems analysis of cultured H9C2 cardiomyoblasts during differentiation over time supports a ‘function follows form’ model of differentiation†
Abstract
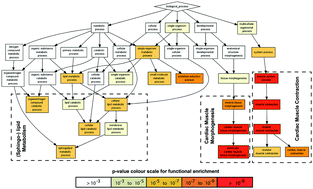
The rat cardiomyoblast cell line H9C2 has emerged as a valuable tool for studying cardiac development, mechanisms of disease and toxicology. We present here a rigorous proteomic analysis that monitored the changes in protein expression during differentiation of H9C2 cells into cardiomyocyte-like cells over time. Quantitative mass spectrometry followed by gene ontology (GO) enrichment analysis revealed that early changes in H9C2 differentiation are related to protein pathways of cardiac muscle morphogenesis and sphingolipid synthesis. These changes in the proteome were followed later in the differentiation time-course by alterations in the expression of proteins involved in cation transport and beta-oxidation. Studying the temporal profile of the H9C2 proteome during differentiation in further detail revealed eight clusters of co-regulated proteins that can be associated with early, late, continuous and transient up- and downregulation. Subsequent reactome pathway analysis based on these eight clusters further corroborated and detailed the results of the GO analysis. Specifically, this analysis confirmed that proteins related to pathways in muscle contraction are upregulated early and transiently, and proteins relevant to extracellular matrix organization are downregulated early. In contrast, upregulation of proteins related to cardiac metabolism occurs at later time points. Finally, independent validation of the proteomics results by immunoblotting confirmed hereto unknown regulators of cardiac structure and ionic metabolism. Our results are consistent with a ‘function follows form’ model of differentiation, whereby early and transient alterations of structural proteins enable subsequent changes that are relevant to the characteristic physiology of cardiomyocytes.


 Please wait while we load your content...
Please wait while we load your content...