Single-gene prognostic signatures for advanced stage serous ovarian cancer based on 1257 patient samples†
Abstract
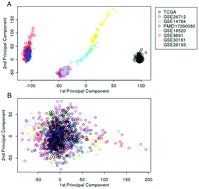
Objective. We sought to identify stable single-gene prognostic signatures based on a large collection of advanced stage serous ovarian cancer (AS-OvCa) gene expression data and explore their functions. Methods. The empirical Bayes (EB) method was used to remove the batch effect and integrate 8 ovarian cancer datasets. Univariate Cox regression was used to evaluate the association between gene and overall survival (OS). The Database for Annotation, Visualization and Integrated Discovery (DAVID) tool was used for the functional annotation of genes for Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Results. The batch effect was removed by the EB method, and 1257 patient samples were used for further analysis. We selected 341 single-gene prognostic signatures with FDR < 0.05, in which 110 and 231 genes were positively and negatively associated with OS, respectively. The functions of these genes were mainly involved in extracellular matrix organization, focal adhesion and DNA replication which are closely associated with cancer. Conclusion. We used the EB method to remove the batch effect of 8 datasets, integrated these datasets and identified stable prognosis signatures for AS-OvCa.


 Please wait while we load your content...
Please wait while we load your content...