Elimination of transforming activity and gene degradation during UV and UV/H2O2 treatment of plasmid-encoded antibiotic resistance genes†
Abstract
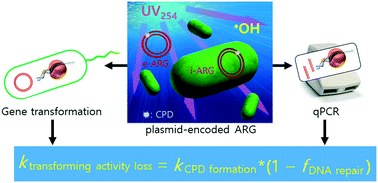
To better understand the elimination of transforming activity of antibiotic resistance genes (ARGs), this study investigated the deactivation of transforming activity of an ARG (in Escherichia coli as a host) and ARG degradation (according to quantitative PCR [qPCR] with different amplicon sizes) during UV (254 nm) and UV/H2O2 treatments of plasmid pUC19 containing an ampicillin resistance gene (ampR). The required UV fluence for each log10 reduction of the transforming activity during UV treatment was ∼37 mJ cm−2 for both extra- and intra-cellular pUC19 (the latter within E. coli). The resulting fluence-based rate constant (k) of ∼6.2 × 10−2 cm2 mJ−1 was comparable to the k determined previously for transforming activity loss of plasmids using host cells capable of DNA repair, but much lower (∼10-fold) than that for DNA repair-deficient cells. The k value for pUC19 transforming activity loss was similarly much lower than the k calculated for cyclobutane-pyrimidine dimer (CPD) formation in the entire plasmid. These results indicate the significant role of CPD repair in the host cells. The degradation rate constants (k) of ampR measured by qPCR increased with increasing target amplicon size (192–851 bp) and were close to the k calculated for the CPD formation in the given amplicons. Further analysis of the degradation kinetics of plasmid-encoded genes from this study and from the literature revealed that qPCR detected most UV-induced DNA damage. In the extracellular plasmid, DNA damage mechanisms other than CPD formation (e.g., base oxidation) were detectable by qPCR and gel electrophoresis, especially during UV/H2O2 treatment. Nevertheless, the enhanced DNA damage for the extracellular plasmids did not result in faster elimination of the transforming activity. Our results indicate that calculated CPD formation rates and qPCR analyses are useful for predicting and/or measuring the rate of DNA damage and predicting the efficiency of transforming activity elimination for plasmid-encoded ARGs during UV-based water disinfection and oxidation processes.
- This article is part of the themed collection: Ultraviolet-based Advanced Oxidation Processes (UV AOPs)


 Please wait while we load your content...
Please wait while we load your content...
