Microbial community structure with trends in methylation gene diversity and abundance in mercury-contaminated rice paddy soils in Guizhou, China†‡
Abstract
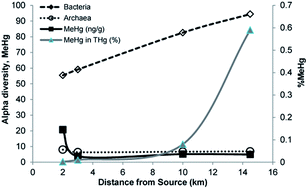
Paddy soils from mercury (Hg)-contaminated rice fields in Guizhou, China were studied with respect to total mercury (THg) and methylmercury (MeHg) concentrations as well as Bacterial and Archaeal community composition. Total Hg (0.25–990 μg g−1) and MeHg (1.3–30.5 ng g−1) varied between samples. Pyrosequencing (454 FLX) of the hypervariable v1–v3 regions of the 16S rRNA genes showed that Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Euryarchaeota, and Crenarchaeota were dominant in all samples. The Bacterial α-diversity was higher in samples with relatively Low THg and MeHg and decreased with increasing THg and MeHg concentrations. In contrast, Archaeal α-diversity increased with increasing of MeHg concentrations but did not correlate with changes in THg concentrations. Overall, the methylation gene hgcAB copy number increased with both increasing THg and MeHg concentrations. The microbial communities at High THg and High MeHg appear to be adapted by species that are both Hg resistant and carry hgcAB genes for MeHg production. The relatively high abundance of both sulfate-reducing δ-Proteobacteria and methanogenic Archaea, as well as their positive correlations with increasing THg and MeHg concentrations, suggests that these microorganisms are the primary Hg-methylators in the rice paddy soils in Guizhou, China.
- This article is part of the themed collection: Mercury Biogeochemistry, Exposure, and Impacts


 Please wait while we load your content...
Please wait while we load your content...
