The effects of implicit modeling of nonpolar solvation on protein folding simulations†
Abstract
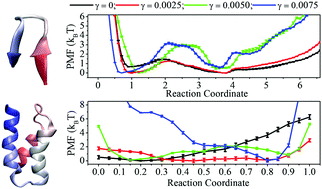
Implicit solvent models, in which the polar and nonpolar solvation free-energies of solute molecules are treated separately, have been widely adopted for molecular dynamics simulation of protein folding. While the development of the implicit models is mainly focused on the methodological improvement and key parameter optimization for polar solvation, nonpolar solvation has been either ignored or described by a simplistic surface area (SA) model. In this work, we performed the folding simulations of multiple β-hairpin and α-helical proteins with varied surface tension coefficients embedded in the SA model to clearly demonstrate the effects of nonpolar solvation treated by a popular SA model on protein folding. The results indicate that the change in the surface tension coefficient does not alter the ability of implicit solvent simulations to reproduce a protein native structure but indeed controls the components of the equilibrium conformational ensemble and modifies the energetic characterization of the folding transition pathway. The suitably set surface tension coefficient can yield explicit solvent simulations and/or experimentally suggested folding mechanism of protein. In addition, the implicit treatment of both polar and nonpolar components of solvation free-energy contributes to the overestimation of the secondary structure in implicit solvent simulations.


 Please wait while we load your content...
Please wait while we load your content...