All-atom molecular dynamics simulations of spin labelled double and single-strand DNA for EPR studies†
Abstract
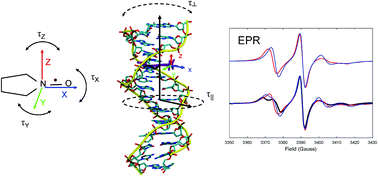
We report the first application of fully atomistic molecular dynamics (MD) simulations to the prediction of electron paramagnetic resonance (EPR) spectra of spin labelled DNA. Models for two structurally different DNA spin probes with either the rigid or flexible position of the nitroxide group in the base pair, employed in experimental studies previously, have been developed. By the application of the combined MD-EPR simulation methodology we aimed at the following. Firstly, to provide a test bed against a sensitive spectroscopic technique for the recently developed improved version of the parmbsc1 force field for MD modelling of DNA. The predicted EPR spectra show good agreement with the experimental ones available from the literature, thus confirming the accuracy of the currently employed DNA force fields. Secondly, to provide a quantitative interpretation of the motional contributions into the dynamics of spin probes in both duplex and single-strand DNA fragments and to analyse their perturbing effects on the local DNA structure. Finally, a combination of MD and EPR allowed us to test the validity of the application of the Model-Free (M-F) approach coupled with the partial averaging of magnetic tensors to the simulation of EPR spectra of DNA systems by comparing the resultant EPR spectra with those simulated directly from MD trajectories. The advantage of the M-F based EPR simulation approach over the direct propagation techniques is that it requires motional and order parameters that can be calculated from shorter MD trajectories. The reported MD-EPR methodology is transferable to the prediction and interpretation of EPR spectra of higher order DNA structures with novel types of spin labels.


 Please wait while we load your content...
Please wait while we load your content...