Efficient structural elucidation of microhydrated biomolecules through the interrogation of hydrogen bond networks†
Abstract
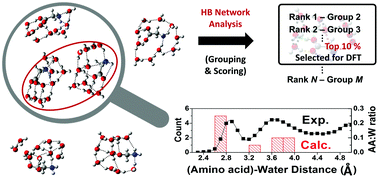
Microhydration of biomolecules is an important structural factor associated with their biological processes. However, there is no general way to elucidate stable hydrated structures even for simple amino acids because of the high complexity of chemical space increasing rapidly with the number of water molecules. Here, we propose a very efficient computational method to selectively sample the most stable structures of microhydrated molecules. The key idea is to utilize the unique structural patterns of H-bond networks obtained from their energetic features, i.e. their tendency to form more H-bonds. As a proof of concept, we could identify the new global minima of glycine·10(H2O) and for the first time, we found the minimum number of water molecules required to stabilize the zwitterionic form of tyrosine. Furthermore, the most stable structures of hydrated glycine and tyrosine indeed had common features, which were consistent with the X-ray data of proteins in water.


 Please wait while we load your content...
Please wait while we load your content...