Binding affinity prediction of nanobody–protein complexes by scoring of molecular dynamics trajectories†
Abstract
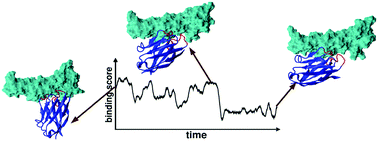
Nanobodies offer a viable alternative to antibodies for engineering high affinity binders. Their small size has an additional advantage: it allows exploiting computational protocols for optimizing their biophysical features, such as the binding affinity. The efficient prediction of this quantity is still considered a daunting task especially for modelled complexes. We show how molecular dynamics can successfully assist in the binding affinity prediction of modelled nanobody–protein complexes. The approximate initial configurations obtained by in silico design must undergo large rearrangements before achieving a stable conformation, in which the binding affinity can be meaningfully estimated. The scoring functions developed for the affinity evaluation of crystal structures will provide accurate estimates for modelled binding complexes if the scores are averaged over long finite temperature molecular dynamics simulations.
- This article is part of the themed collection: 2018 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...