Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches†
Abstract
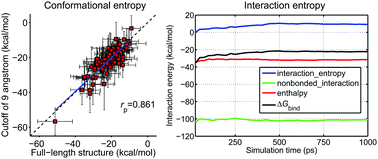
Entropy effects play an important role in drug–target interactions, but the entropic contribution to ligand-binding affinity is often neglected by end-point binding free energy calculation methods, such as MM/GBSA and MM/PBSA, due to the expensive computational cost of normal mode analysis (NMA). Here, we systematically investigated entropy effects on the prediction power of MM/GBSA and MM/PBSA using >1500 protein–ligand systems and six representative AMBER force fields. Two computationally efficient methods, including NMA based on truncated structures and the interaction entropy approach, were used to estimate the entropic contributions to ligand–target binding free energies. In terms of the overall accuracy, we found that, for the minimized structures, in most cases the inclusion of the conformational entropies predicted by truncated NMA (enthalpynmode_min_9Å) compromises the overall accuracy of MM/GBSA and MM/PBSA compared with the enthalpies calculated based on the minimized structures (enthalpymin). However, for the MD trajectories, the binding free energies can be improved by the inclusion of the conformation entropies predicted by either truncated-NMA for a relatively high dielectric constant (εin = 4) or the interaction entropy method for εin = 1–4. In terms of reproducing the absolute binding free energies, the binding free energies estimated by including the truncated-NMA entropies based on the MD trajectories (ΔGnmode_md_9Å) give the lowest average absolute deviations against the experimental data among all the tested strategies for both MM/GBSA and MM/PBSA. Although the inclusion of the truncated NMA based on the MD trajectories (ΔGnmode_md_9Å) for a relatively high dielectric constant gave the overall best result and the lowest average absolute deviations against the experimental data (for the ff03 force field), it needs too much computational time. Alternatively, considering that the interaction entropy method does not incur any additional computational cost and can give comparable (at high dielectric constant, εin = 4) or even better (at low dielectric constant, εin = 1–2) results than the truncated-NMA entropy (ΔGnmode_md_9Å), the interaction entropy approach is recommended to estimate the entropic component for MM/GBSA and MM/PBSA based on MD trajectories, especially for a diverse dataset. Furthermore, we compared the predictions of MM/GBSA with six different AMBER force fields. The results show that the ff03 force field (ff03 for proteins and gaff with AM1-BCC charges for ligands) performs the best, but the predictions given by the tested force fields are comparable, implying that the MM/GBSA predictions are not very sensitive to force fields.
- This article is part of the themed collection: 2018 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...