Tuning protein assembly pathways through superfast amyloid-like aggregation†
Abstract
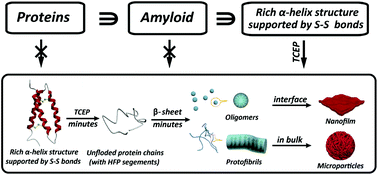
Amyloid formation of proteins is not only relevant for neurodegenerative diseases, but has recently emerged as a groundbreaking approach in materials science and biotechnology. However, amyloid aggregation of proteins in vitro generally requires a long incubation time under extremely harsh conditions, and the understanding of the structural motif to determine amyloid assembly is extremely limited. Herein we reveal that the integration of three important building blocks in typical globular proteins is crucial for superfast protein amyloid-like assembly including the segment required for high fibrillation propensity, abundant α-helix structures and intramolecular S–S bonds to lock the α-helix. With the reduction of the S–S bond by tris(2-carboxyethyl)phosphine (TCEP), the α-helix was rapidly unlocked from the protein chain, and the resultant unfolded monomer underwent a fast transition to β-sheet-rich amyloid oligomers and protofibrils in minutes, which further assembled into a macroscopic nanofilm at the air/water interface and microparticles in bulk solution, respectively.
- This article is part of the themed collection: Biomaterials Science 10th Anniversary: Top papers from China


 Please wait while we load your content...
Please wait while we load your content...