Regional feature extraction of various fishes based on chemical and microbial variable selection using machine learning†
Abstract
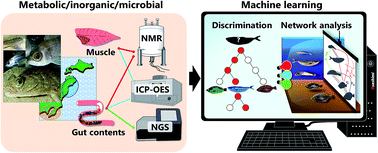
We introduce a method for extracting regional and habitat features of various fish species based on chemical and microbial correlations that incorporate integrated analysis and a variable selection approach. We characterized 24 fish species from two marine regions in Japan, in terms of the metabolic and inorganic profiles of muscle and gut contents, as well as gut microbes. Using machine learning, the integrated analysis based on the metabolic, inorganic, and microbial profiles of muscle and gut contents allows the characterization of both the fish species and habitat regions. The results revealed that the fish muscle tissue profile provides high-value data for evaluating ecosystems and discriminating fish populations based on species and regions. To visualize the regionality and habitat, we developed a method to efficiently extract the most important variables using the machine learning approach, followed by correlation analysis of variations in muscle and gut content profiles. The correlation networks enabled efficient visualization of marine ecosystems in the Tohoku and Kanto regions of Japan. This method should be useful for evaluating fish habitats and elucidating associated environmental chemical networks.
- This article is part of the themed collection: Analytical Methods Recent Open Access Articles


 Please wait while we load your content...
Please wait while we load your content...