Engineering oligonucleotide-based peroxidase mimetics for the colorimetric assay of S1 nuclease†
Abstract
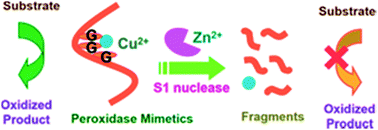
Herein we propose a rational design to construct peroxidase mimetics in response to S1 nuclease by employing guanine-rich oligonucleotides and Cu2+ ions. The enzymatic activities of DNA–Cu(II) are highly dependent upon the binding stoichiometry between Cu2+ and DNA. In the H2O2-mediated oxidation of 3,3′,5,5′-tetramethylbenzidine, the catalytic activity of Cu2+ can be accelerated approximately 88 times in the presence of AG3(T2AG3)3, whereas it shows slight enhancement in the presence of a mononucleotide (GMP). The coordination of Cu2+ with DNA structures greatly strengthens its ability to generate reactive oxygen species. The most intriguing finding in this study is that DNA–Cu(II) complexes lose their enzymatic activities after the cleavage of DNA scaffolds by S1 nuclease. A colorimetric method was established for selectively evaluating the S1 nuclease activity with a limit of detection of 4.7 × 10−3 U mL−1, only by using label-free oligonucleotides and copper ions without complicated synthesis and apparatus. This facile method can also be applicable for quantitative analysis in biological fluids.
- This article is part of the themed collection: Analytical Methods Recent HOT articles


 Please wait while we load your content...
Please wait while we load your content...