Deducing disulfide patterns of cysteine-rich proteins using signature fragments produced by top-down mass spectrometry†
Abstract
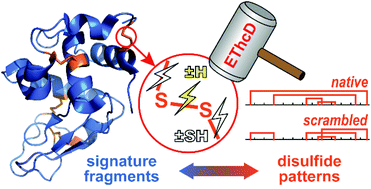
Direct mapping of protein disulfide patterns using top-down mass spectrometry (MS) is often hampered by inadequate fragmentation at the disulfide-enclosing region, and insufficient structural information provided by the fragments. Here we used electron-transfer/high energy collision dissociation (EThcD) to improve the fragmentation efficiency, and developed strategies that minimize the false positive identification of fragments and deconvolute the signals representing specific modifications made to the disulfide-cleavage-induced fragments. We observed clear correlations between unique modification (attachment or removal of H or SH) patterns and the number of disulfide bonds that enclose the corresponding region. Using the characteristic signature fragments, we in part localized the Cys-bridging sites in disulfide-scrambled lysozymes, and reduced the number of putative disulfide patterns from 104 to 6. The results demonstrated the feasibility of direct analysis of complex disulfide patterns using top-down MS.
- This article is part of the themed collection: Analyst Recent HOT articles


 Please wait while we load your content...
Please wait while we load your content...