Simple and rapid detection of bacteria using a nuclease-responsive DNA probe†
Abstract
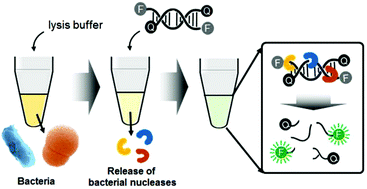
We demonstrate simple and rapid bacterial detection using a nuclease-responsive DNA probe. The probe consisting of a fluorescent dye and a quencher at the 5′ and 3′ termini, respectively, was designed to be cleaved by nucleases such as endonucleases, exonucleases, and DNases, which are released from bacteria using an optimized lysis buffer. The fluorescence signal of the cleaved DNA probe correlates with the number of Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus, and the detection limit was 103 CFU for E. coli and 104 CFU for S. aureus. Moreover, this method is specific for live bacteria and takes just one minute to get the signal including sample collection. These features make the present bacterial detection method a powerful on-site bacterial contamination assay which is simple, rapid, and quantitative.


 Please wait while we load your content...
Please wait while we load your content...