Analyzing the structure of macromolecules in their native cellular environment using hydroxyl radical footprinting
Abstract
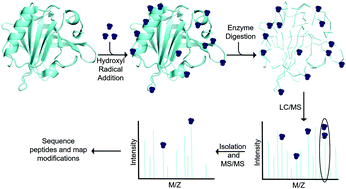
Hydroxyl radical footprinting (HRF) has been successfully used to study the structure of both nucleic acids and proteins. The method utilizes hydroxyl radicals to oxidize solvent accessible sites in macromolecules. In recent years, the method has shown some utility for live cell analysis. In this review, we will survey the current state of the field for footprinting macromolecules in living cells. The field is relatively new, particularly for protein studies, with only a few publications on the development and application of HRF on live cells. DNA–protein interaction sites and information on the secondary and tertiary structure of RNA has been characterized. In addition, the conformational changes of membrane-spanning channels upon opening and activation have also been studied by in-cell HRF. In this review, we highlight examples of these applications.
- This article is part of the themed collection: Recent Review Articles


 Please wait while we load your content...
Please wait while we load your content...