A simplified characterization of S-adenosyl-l-methionine-consuming enzymes with 1-Step EZ-MTase: a universal and straightforward coupled-assay for in vitro and in vivo setting†
Abstract
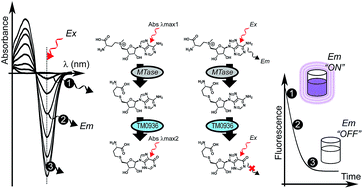
Methyltransferases use S-adenosyl-L-methionine (SAM) to deposit methyl marks. Many of these epigenetic ‘writers’ are associated with gene regulation. As cancer etiology is highly correlated with misregulated methylation patterns, methyltransferases are emerging therapeutic targets. Successful assignment of methyltransferases' roles within intricate biological networks relies on (1) the access to enzyme mechanistic insights and (2) the efficient screening of chemical probes against these targets. To characterize methyltransferases in vitro and in vivo, we report a highly-sensitive one-step deaminase-linked continuous assay where the S-adenosyl-L-homocysteine (SAH) enzyme-product is rapidly and quantitatively catabolized to S-inosyl-L-homocysteine (SIH). To highlight the broad capabilities of this assay, we established enzymatic characteristics of two protein arginine methyltransferases (PRMT5 and PRMT7), a histone-lysine N-methyltransferase (DIM-5) and a sarcosine/dimethylglycine N-methyltransferase (SDMT). Since the coupling deaminase TM0936 displays robust activity over a broad pH-range we determined the pH dependence of SDMT reaction rates. TM0936 reactions are monitored at 263 nm, so a drawback may arise when methyl acceptor substrates absorb within this UV-range. To overcome this limitation, we used an isosteric fluorescent SAM-analog: S-8-aza-adenosyl-L-methionine. Most enzymes tolerated this probe and sustained methyltransfers were efficiently monitored through loss of fluorescence at 360 nm. Unlike discontinuous radioactive- and antibody-based assays, our assay provides a simple, versatile and affordable approach towards the characterization of methyltransferases. Supported by three logs of linear dynamic range, the 1-Step EZ-MTase can detect methylation rates as low as 2 μM h−1, thus making it possible to quantify low nanomolar concentrations of glycine N-methyltransferase within crude biological samples. With Z′-factors above 0.75, this assay is well suited to high-throughput screening and may promote the identification of novel therapeutics.


 Please wait while we load your content...
Please wait while we load your content...