Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination–amplification strategy†
Abstract
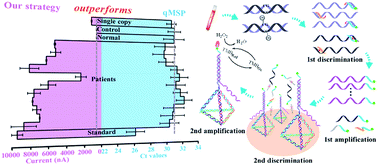
Tumor-related circulating methylated DNA represents only a small fraction of the total DNA in clinical samples (e.g. plasma), challenging the accurate analysis of specific DNA methylation patterns. Yet conventional assays based on the real-time quantitative methylation-specific PCR (qMSP) are generally limited in detection sensitivity and specificity due to its non-specific amplification interference including primer dimers and off-target amplification. Here we propose a single copy-sensitive electrochemical assay for circulating methylated DNA with ultrahigh specificity on the basis of a sequential discrimination–amplification strategy. Methylated DNA rather than unmethylated DNA in a bisulfite-modified sample is identified and amplified by the asymmetric MSP to generate abundant biotin-labeled single-stranded amplicons with reduced primer–dimer artifacts. Self-assembled tetrahedral DNA probes, which are readily decorated on an electrode surface as nanostructured probes with ordered orientation and well controlled spacing, enable the highly efficient hybridization of the specific single-stranded amplicons due to greatly increased target accessibility and significantly decreased noise. The interfacial hybridization event is quantitatively translated into electrochemical signals utilizing an enzymatic amplification. The proposed assay integrates dual sequence discrimination processes and cascade signal amplification processes, achieving the identification of as few as one methylated DNA molecule in the presence of a 1000-fold excess of unmethylated alleles. Furthermore, the excellent assay performance enables tumor related methylation detection in lung cancer patients with 200 microlitre plasma samples. The results are in good consistency with those of clinical diagnosis, whereas the conventional qMSP failed to detect the corresponding methylation pattern of these clinically confirmed positive patients in such trace amounts of samples.


 Please wait while we load your content...
Please wait while we load your content...