Mapping a protein recognition centre with chiral photoactive ligands. An integrated approach combining photophysics, reactivity, proteomics and molecular dynamics simulation studies†
Abstract
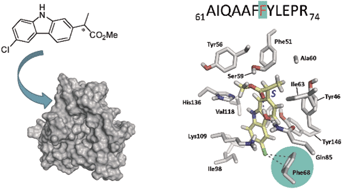
A multidisciplinary strategy to obtain structural information on the intraprotein region is described here. As probe ligands, (S)- and (R)-CPFMe (the methyl esters of the chiral drug carprofen) have been selected, while bovine α1-acid glycoprotein (BAAG) has been chosen as a biological host. The procedure involves the separate irradiation of the BAAG/(S)-CPFMe and BAAG/(R)-CPFMe complexes, coupled with fluorescence spectroscopy, laser flash photolysis, proteomic analysis, docking and molecular dynamics simulations. Thus, irradiation of the BAAG/CPFMe complexes at λ = 320 nm was followed by fluorescence spectroscopy. The intensity of the emission band obtained after irradiation indicated photodehalogenation, whereas its structureless shape suggested covalent binding of the resulting radical CBZMe˙ to the biopolymer. After gel filtration chromatography, the spectra still displayed emission, in agreement with covalent attachment of CBZMe˙ to BAAG. Stereodifferentiation was observed in this process. After trypsin digestion and ESI-MS/MS, the incorporation of CBZMe was detected at Phe68. Docking and molecular dynamics simulation studies, which were carried out using a homology model of BAAG, reveal that the closer proximity of the aromatic moiety of the (S)-enantiomer to the phenyl group of Phe68 would be responsible for the experimentally observed, more effective chemical modification of the protein. The proposed tridimensional structure of BAAG covalently modified by the two enantiomers is also provided. In principle, this approach can be extended to a variety of protein/ligand complexes.


 Please wait while we load your content...
Please wait while we load your content...