Effect of polymer topology on non-covalent polymer–protein complexation: miktoarm versus linear mPEG-poly(glutamic acid) copolymers†
Abstract
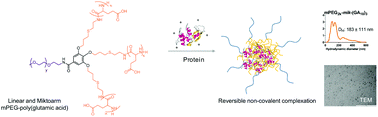
Non-covalent polymer–protein conjugation is emerging as a potential route to improve pharmacokinetics and pharmacodynamics of protein therapeutics. In this study, a family of structurally related block copolymers of mPEG2k – poly(glutamic acid) with linear A-B (mPEG2k-lin-polyGA) and miktoarm A-B3 (mPEG2k-mik-(polyGA)3) structure was synthesised by N-carboxyanhydride (NCA) ring-opening polymerisation to assess the effect of macromolecular topology of the copolymers on polymer–protein complexation. The data illustrate that the synthesised copolymers are capable of complexing a model protein, lysozyme, at optimal pH conditions through non-covalent interactions, with complexation efficiencies depending on the copolymers composition and molecular architecture. In native gel electrophoresis experiments, linear mPEG2k-lin-GA10 copolymer, possessing a short polyanionic polyGA block, shows a low level of complexation, which does not change when the number of polyGA branches of the same size is increased, using a miktoarm mPEG2k-mik-(GA10)3 copolymer. However, enhanced complexation is observed when the same number of ionisable GA units (30) are displayed on a linear macromolecular scaffold; mPEG2k-mik-(GA10)3vs. mPEG2k-lin-GA30. Again complexation efficiency did not increase when the number of complexing polyGA branches were increased; mPEG2k-lin-GA30 vs. mPEG2k-mik-(GA30)3. Nanoparticle tracking analysis (NTA) showed that the copolymer–protein complexes possessed hydrodynamic diameters in the 50–200 nm range, suggesting a degree of control in the assembly process. Sequestration of lysozyme within polymer complexes resulted in a decrease in its apparent enzymatic activity, which was re-established on the complexes dissociation upon a treatment with competitive complexant. Intrinsic fluorescence and circular dichroism (CD) studies suggested structural conformation of the protein was not altered following complexation with mPEG2k-polyGA copolymers. Taken together, these results provide an initial structure–function relationship for protein-complexing mPEG2k-polyGA copolymers with variable macromolecular topology, opening the way for their future application in biological and biomedical studies.


 Please wait while we load your content...
Please wait while we load your content...