Molecular simulations of peptide amphiphiles
Abstract
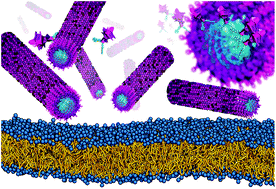
This review describes recent progress in the area of molecular simulations of peptide assemblies, including peptide-amphiphiles and drug-amphiphiles. The ability to predict the structure and stability of peptide self-assemblies from the molecular level up is vital to the field of nanobiotechnology. Computational methods such as molecular dynamics offer the opportunity to characterize intermolecular forces between peptide-amphiphiles that are critical to the self-assembly process. Furthermore, these computational methods provide the ability to computationally probe the structure of these supramolecular assemblies at the molecular level, which is a challenge experimentally. Herein, we briefly highlight progress in the areas of all-atomistic and coarse-grained simulation studies investigating the self-assembly process of short peptides and peptide amphiphiles. We also discuss recent all-atomistic and coarse-grained simulations of the self-assembly of a drug-amphiphile into elongated filaments. Next, we discuss how these computational methods can provide further insight into the pathway of cylindrical nanofiber formation and predict their biocompatibility by studying the interaction of these peptide-amphiphile nanostructures with model cell membranes.
- This article is part of the themed collection: Peptide Materials


 Please wait while we load your content...
Please wait while we load your content...