Confinement-driven organization of a histone-complexed DNA molecule in a dense array of nanoposts†
Abstract
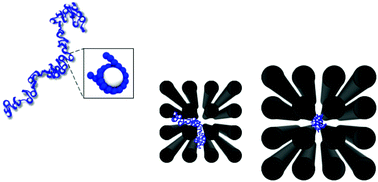
The first step in the controlled storage of lengthy DNA molecules is to keep DNA molecules separated while integrated in micrometer-sized space. Herein, we present hybrid Monte Carlo simulations of a histone-complexed DNA (hcDNA) molecule confined in a dense array of nanoposts. Depending on the nanopost dimension, a single, 8.7 kilobase pair hcDNA molecule was either localized and elongated in a single inter-post space surrounded by four nanoposts or spread over several inter-post spaces through passages between two neighboring nanoposts. The conformational change of a hcDNA molecule is interpreted in terms of competitive effects of confinements in the inter-post and passage spaces. We propose that, by elaborately designing nanopost arrays, the competitive confinement effects can be adjusted such that each hcDNA molecule is localized in a single inter-post space, and thereby multiple hcDNA molecules can be physically separated from each other while stored together in the nanopost array.


 Please wait while we load your content...
Please wait while we load your content...