Gene co-opening network deciphers gene functional relationships†
Abstract
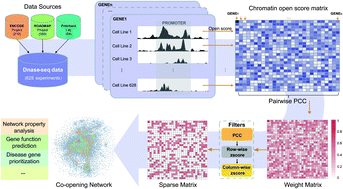
Genome sequencing technology has generated a vast amount of genomic and epigenomic data, and has provided us a great opportunity to study gene functions on a global scale from an epigenomic view. In the last decade, network-based studies, such as those based on PPI networks and co-expression networks, have shown good performance in capturing functional relationships between genes. However, the functions of a gene and the mechanism of interaction of genes with each other to elucidate their functions are still not entirely clear. Here, we construct a gene co-opening network based on chromatin accessibility of genes. We show that genes related to a specific biological process or the same disease tend to be clustered in the co-opening network. This understanding allows us to detect functional clusters from the network and to predict new functions for genes. We further apply the network to prioritize disease genes for Psoriasis, and demonstrate the power of the joint analysis of the co-opening network and GWAS data in identifying disease genes. Taken together, the co-opening network provides a new viewpoint for the elucidation of gene associations and the interpretation of disease mechanisms.


 Please wait while we load your content...
Please wait while we load your content...