Hybrid deterministic/stochastic simulation of complex biochemical systems
Abstract
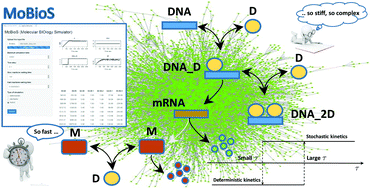
In a biological cell, cellular functions and the genetic regulatory apparatus are implemented and controlled by complex networks of chemical reactions involving genes, proteins, and enzymes. Accurate computational models are indispensable means for understanding the mechanisms behind the evolution of a complex system, not always explored with wet lab experiments. To serve their purpose, computational models, however, should be able to describe and simulate the complexity of a biological system in many of its aspects. Moreover, it should be implemented by efficient algorithms requiring the shortest possible execution time, to avoid enlarging excessively the time elapsing between data analysis and any subsequent experiment. Besides the features of their topological structure, the complexity of biological networks also refers to their dynamics, that is often non-linear and stiff. The stiffness is due to the presence of molecular species whose abundance fluctuates by many orders of magnitude. A fully stochastic simulation of a stiff system is computationally time-expensive. On the other hand, continuous models are less costly, but they fail to capture the stochastic behaviour of small populations of molecular species. We introduce a new efficient hybrid stochastic–deterministic computational model and the software tool MoBioS (MOlecular Biology Simulator) implementing it. The mathematical model of MoBioS uses continuous differential equations to describe the deterministic reactions and a Gillespie-like algorithm to describe the stochastic ones. Unlike the majority of current hybrid methods, the MoBioS algorithm divides the reactions' set into fast reactions, moderate reactions, and slow reactions and implements a hysteresis switching between the stochastic model and the deterministic model. Fast reactions are approximated as continuous-deterministic processes and modelled by deterministic rate equations. Moderate reactions are those whose reaction waiting time is greater than the fast reaction waiting time but smaller than the slow reaction waiting time. A moderate reaction is approximated as a stochastic (deterministic) process if it was classified as a stochastic (deterministic) process at the time at which it crosses the threshold of low (high) waiting time. A Gillespie First Reaction Method is implemented to select and execute the slow reactions. The performances of MoBios were tested on a typical example of hybrid dynamics: that is the DNA transcription regulation. The simulated dynamic profile of the reagents’ abundance and the estimate of the error introduced by the fully deterministic approach were used to evaluate the consistency of the computational model and that of the software tool.


 Please wait while we load your content...
Please wait while we load your content...