Hedgehog-mesenchyme gene signature identifies bi-modal prognosis in luminal and basal breast cancer sub-types†
Abstract
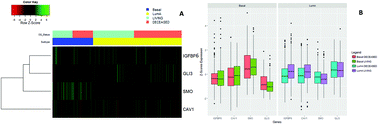
Hedgehog signaling (Hh) has been shown to be hyper-activated in several cancers. However, active Hh signaling can promote or inhibit tumor growth; thus identification of markers beyond main canonical Hh target genes is needed to improve patient selection and clinical outcome in response to Hh inhibitors. Cancer-associated fibroblasts (CAFs) have been linked with tumor progression and beneficial response to Hh inhibitors. Thus, we hypothesized that genes associated with Hh-activated CAFs can be used for stratification of tumors that will benefit from Hh inhibitors. In this work, we evaluated a 15-gene fingerprint that combines Hh and mesenchymal genes associated with CAF phenotype to profile breast cancer sub-types based on gene expression patterns among clustered groups. About 3800 cancer samples were evaluated using random forest models and linear discriminant analysis to sort breast cancer by subtypes and therapeutic approach. The results showed that the Hh-mesenchyme gene fingerprint has a highly sensitive and differential expression pattern among basal and luminal A sub-groups. Basal samples with high levels of Hh target genes had better prognosis than luminal A samples. Luminal A samples with a tendency towards Hh signaling suppression had higher overall and disease-free survival rates particularly if deprived of hormone therapy. Hh transcriptional repressor GLI3 and signaling activator SMO were the top 2 genes for discriminating among samples with active Hh signaling in human breast cancer subtypes and Hh-inhibitor resistant tumors. Caveolin-1 (CAV1), a gene with low expression in CAFs, shows strong correlation with active Hh signaling and discrimination among survival curves in luminal A patients with active or inactive Hh signaling. Our data suggest that CAV1 is an important gene for monitoring Hh inhibition in tumors and support further stratification by hormone therapy status prior to use of Hh inhibitors.


 Please wait while we load your content...
Please wait while we load your content...