LPI-ETSLP: lncRNA–protein interaction prediction using eigenvalue transformation-based semi-supervised link prediction
Abstract
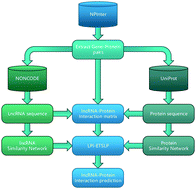
RNA–protein interactions are essential for understanding many important cellular processes. In particular, lncRNA–protein interactions play important roles in post-transcriptional gene regulation, such as splicing, translation, signaling and even the progression of complex diseases. However, the experimental validation of lncRNA–protein interactions remains time-consuming and expensive, and only a few theoretical approaches are available for predicting potential lncRNA–protein associations. Here, we presented eigenvalue transformation-based semi-supervised link prediction (LPI-ETSLP) to uncover the relationship between lncRNAs and proteins. Moreover, it is semi-supervised and does not need negative samples. Based on 5-fold cross validation, an AUC of 0.8876 and an AUPR of 0.6438 have demonstrated its reliable performance compared with three other computational models. Furthermore, the case study demonstrated that many lncRNA–protein interactions predicted by our method can be successfully confirmed by experiments. It is indicated that LPI-ETSLP would be a useful bioinformatics resource for biomedical research studies.


 Please wait while we load your content...
Please wait while we load your content...