An integrative machine learning strategy for improved prediction of essential genes in Escherichia coli metabolism using flux-coupled features†
Abstract
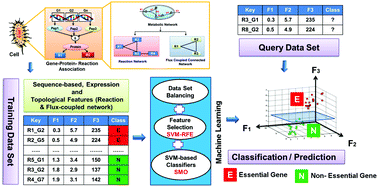
Prediction of essential genes helps to identify a minimal set of genes that are absolutely required for the appropriate functioning and survival of a cell. The available machine learning techniques for essential gene prediction have inherent problems, like imbalanced provision of training datasets, biased choice of the best model for a given balanced dataset, choice of a complex machine learning algorithm, and data-based automated selection of biologically relevant features for classification. Here, we propose a simple support vector machine-based learning strategy for the prediction of essential genes in Escherichia coli K-12 MG1655 metabolism that integrates a non-conventional combination of an appropriate sample balanced training set, a unique organism-specific genotype, phenotype attributes that characterize essential genes, and optimal parameters of the learning algorithm to generate the best machine learning model (the model with the highest accuracy among all the models trained for different sample training sets). For the first time, we also introduce flux-coupled metabolic subnetwork-based features for enhancing the classification performance. Our strategy proves to be superior as compared to previous SVM-based strategies in obtaining a biologically relevant classification of genes with high sensitivity and specificity. This methodology was also trained with datasets of other recent supervised classification techniques for essential gene classification and tested using reported test datasets. The testing accuracy was always high as compared to the known techniques, proving that our method outperforms known methods. Observations from our study indicate that essential genes are conserved among homologous bacterial species, demonstrate high codon usage bias, GC content and gene expression, and predominantly possess a tendency to form physiological flux modules in metabolism.


 Please wait while we load your content...
Please wait while we load your content...