FOCuS: a metaheuristic algorithm for computing knockouts from genome-scale models for strain optimization†
Abstract
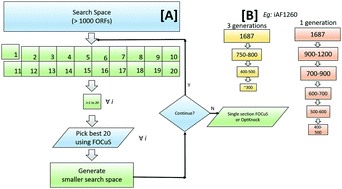
Although handful tools are available for constraint-based flux analysis to generate knockout strains, most of these are either based on bilevel-MIP or its modifications. However, metaheuristic approaches that are known for their flexibility and scalability have been less studied. Moreover, in the existing tools, sectioning of search space to find optimal knocks has not been considered. Herein, a novel computational procedure, termed as FOCuS (Flower-pOllination coupled Clonal Selection algorithm), was developed to find the optimal reaction knockouts from a metabolic network to maximize the production of specific metabolites. FOCuS derives its benefits from nature-inspired flower pollination algorithm and artificial immune system-inspired clonal selection algorithm to converge to an optimal solution. To evaluate the performance of FOCuS, reported results obtained from both MIP and other metaheuristic-based tools were compared in selected case studies. The results demonstrated the robustness of FOCuS irrespective of the size of metabolic network and number of knockouts. Moreover, sectioning of search space coupled with pooling of priority reactions based on their contribution to objective function for generating smaller search space significantly reduced the computational time.


 Please wait while we load your content...
Please wait while we load your content...