Optimal parameter values for the control of gene regulation†
Abstract
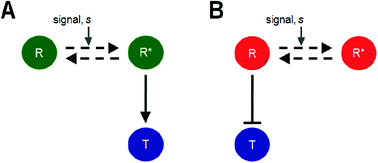
How does a transcription network arrive at the particular values of biochemical interactions defining it? These interactions define DNA-transcription factor interaction, degradation rates of proteins, promoter strengths, and communication of the environmental signal with the network. What is the structure of the fitness landscape that is defined by the space that these parameters can take on? To answer these questions, we simulate the simplest regulatory network: a transcription factor, R, and a target protein, T. We use a cost-benefit analysis to evolve the network and eventually arrive at values of parameters which maximize fitness. We show that for a given topology, multiple parameter sets exist which confer maximal fitness to the cell, and that pairwise correlations exist between parameters in optimal sets. In addition, our results indicate that in the parameter space defining the interactions in a topology, a highly rugged fitness landscape exists.


 Please wait while we load your content...
Please wait while we load your content...