Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics
Abstract
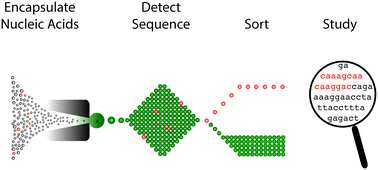
Nucleic acids encode the information of life, programming cellular functions and dictating many biological outcomes. Differentiating between cells based on their nucleic acid programs is, thus, a powerful way to unravel the genetic bases of many phenotypes. This is especially important considering that most cells exist in heterogeneous populations, requiring them to be isolated before they can be studied. Existing flow cytometry techniques, however, are unable to reliably recover specific cells based on nucleic acid content. Nucleic acid cytometry is a new field built on droplet microfluidics that allows robust identification, sorting, and sequencing of cells based on specific nucleic acid biomarkers. This review highlights applications that immediately benefit from the approach, biological questions that can be addressed for the first time with it, and considerations for building successful workflows.
- This article is part of the themed collections: Lab on a Chip Recent Review Articles and Droplet-based single-cell sequencing


 Please wait while we load your content...
Please wait while we load your content...