Microscopic understanding of the conformational features of a protein–DNA complex†
Abstract
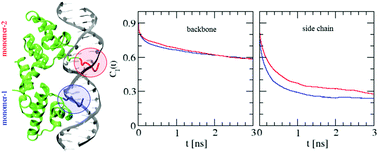
Protein–DNA interactions play crucial roles in different biological processes. Binding of a protein to its target DNA is the key step at different stages of genetic activities. In this article, we have carried out atomistic molecular dynamics simulations to understand the microscopic conformational and dynamical features of the N-terminal domain of the λ-repressor protein and its operator DNA in their complexed state. The calculations revealed that the overall flexibility of the protein and the DNA components reduces due to complex formation. In particular, increased ordering of the DNA sugar rings bound to the protein is found to be associated with modified ring puckering. Attempts have been made to study the effect of complexation on the internal motions of the protein and the DNA components. It is demonstrated that the non-uniform ordering of the side chains of lysine residues in the consensus sequence leads to differential behavior of the two monomers of the homodimeric protein.


 Please wait while we load your content...
Please wait while we load your content...