Enzymatic activity inside a DNA/peptide complex†
Abstract
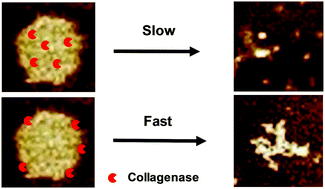
The mutual interaction between enzymes and their environments plays a key role in various life processes. In this study, using the complexes formed by salmon DNA and a de novo designed peptide, Ac-RRRRRRRRRGALGLPGKGGGLQRLTALDGR-NH2 (abbreviated as RR-30), as a model, we studied the activity of collagenase encapsulated inside the complex. Collagenase is able to cleave RR-30 at a LG/LP site, generating two shorter length peptides, which decreases the stability of the complex. Results show that the complex dissociates with time in the presence of collagenase. The dissociation rate is linearly proportional to the collagenase concentration. On the other hand, the collagenase activity is severely deteriorated inside the complex, where only 1/3 of the enzyme is active. We attribute it to the electrostatic interaction and hydrophobic interaction between collagenase and the components of the complex. Therefore, the mutual interaction determines the structure and kinetics of the DNA/peptide complex.


 Please wait while we load your content...
Please wait while we load your content...