Toward a quantitative description of microscopic pathway heterogeneity in protein folding†
Abstract
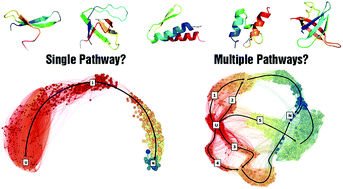
How many structurally different microscopic routes are accessible to a protein molecule while folding? This has been a challenging question to address experimentally as single-molecule studies are constrained by the limited number of observed folding events while ensemble measurements, by definition, report only an average and not the distribution of the quantity under study. Atomistic simulations, on the other hand, are restricted by sampling and the inability to reproduce thermodynamic observables directly. We overcome these bottlenecks in the current work and provide a quantitative description of folding pathway heterogeneity by developing a comprehensive, scalable and yet experimentally consistent approach combining concepts from statistical mechanics, physical kinetics and graph theory. We quantify the folding pathway heterogeneity of five single-domain proteins under two thermodynamic conditions from an analysis of 100 000 folding events generated from a statistical mechanical model incorporating the detailed energetics from more than a million conformational states. The resulting microstate energetics predicts the results of protein engineering experiments, the thermodynamic stabilities of secondary-structure segments from NMR studies, and the end-to-end distance estimates from single-molecule force spectroscopy measurements. We find that a minimum of ∼3–200 microscopic routes, with a diverse ensemble of transition-path structures, are required to account for the total folding flux across the five proteins and the thermodynamic conditions. The partitioning of flux amongst the numerous pathways is shown to be subtly dependent on the experimental conditions that modulate protein stability, topological complexity and the structural resolution at which the folding events are observed. Our predictive methodology thus reveals the presence of rich ensembles of folding mechanisms that are generally invisible in experiments, reconciles the contradictory observations from experiments and simulations and provides an experimentally consistent avenue to quantify folding heterogeneity.


 Please wait while we load your content...
Please wait while we load your content...