On the role of topology in regulating transcriptional cascades†
Abstract
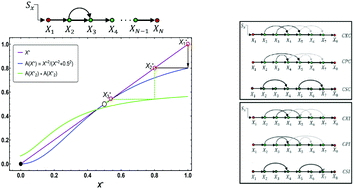
We study the impact of topology on the response of a transcriptional cascade with certain circuit topologies to a constant and time-varying input signal. We systematically analyze the response of the output to activating and repressing cascades. We identify two types of responses for a linear cascade, namely the “Decaying mode”, where the input signal becomes exceedingly weaker as it propagates, and the “Bistable mode”, where the input signal can either be amplified or die out in the pathway. We examine how the transition occurs from one mode to the other as we add coherent and/or incoherent feed-forward loops in an otherwise linear cascade. We find that pathways with at least one incoherent feedforward loop can perform adaptive responses with the quality of response varying among different topologies. Furthermore, we study the origin of a (non)monotonic input–output profile for various circuit topologies over a wide range of parameter space. For a time-varying input signal, we identify some circuit topologies that are more prone to noise propagation than others that are more reliable in blocking out high-amplitude fluctuations. We discuss the effect of cell to cell variation in protein expression on the output of a linear cascade and compare the robustness of activating and repressing cascades to noise propagations. In the end, we apply our model to study an example of a transcription cascade that guides the development of Bacillus subtilis spores and discuss an example from a metabolic pathway where a transition from the decaying to bistable mode can occur by changing the topology of interactions in the pathway.


 Please wait while we load your content...
Please wait while we load your content...