Structural and dynamical instability of DNA caused by high occurrence of d5SICS and dNaM unnatural nucleotides†
Abstract
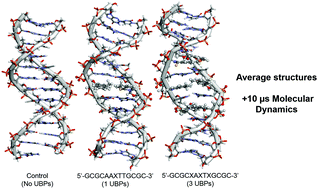
In vivo inclusion of unnatural base pairs (UBPs) into functional DNA was recently reported for compounds 2,6-dimethyl-2H-isoquiniline-1-thione (d5SICS, X) and 2-methoxy-3-methylnaphthalene (dNaM, Y) in a modified E. coli strand, for which high-fidelity replication was observed. Yet, little is known about possible genetic consequences they have in largely substituted DNA. Using a converged microsecond long molecular dynamics (MD) simulation of the sequences 5′-GCGCAAXTTGCGC-3′ and 5′-GCGCXAXTXGCGC-3′, where X represents the UBP, we show that in the system with only a single XY UBP pair present, the global RMS deviation from canonical B-DNA in our control simulations is ∼3 Å and a fully converged ensemble is achieved within 2 µs. With three UBPs, deviation increases to ∼5 Å and convergence is not achieved within 10 µs of sampling time. With five UBPs, no convergence is observed and the double helix collapses into a globular structure. A fully optimized structure of the trimer d(GXC) was obtained using the density functional theory method B97D/cc-pVTZ and resulted in an RMSD value of ∼2 Å when compared to the most populated structure obtained from the MD simulations. Their calculated interaction energy is −3.7 kcal mol−1. It is thus unlikely that d5SICS and dNaM could be useful as tools in DNA manipulation. This theoretical methodology can be used in the de novo design of UBPs.


 Please wait while we load your content...
Please wait while we load your content...